A tripartite rheostat controls self-regulated host plant resistance to insects
- PMID: 37316670
- PMCID: PMC10284691
- DOI: 10.1038/s41586-023-06197-z
A tripartite rheostat controls self-regulated host plant resistance to insects
Abstract
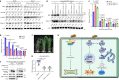
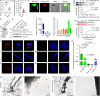
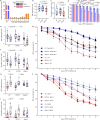
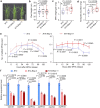
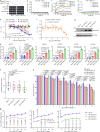
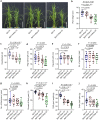
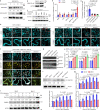
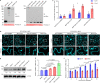
Plants deploy receptor-like kinases and nucleotide-binding leucine-rich repeat receptors to confer host plant resistance (HPR) to herbivores1. These gene-for-gene interactions between insects and their hosts have been proposed for more than 50 years2. However, the molecular and cellular mechanisms that underlie HPR have been elusive, as the identity and sensing mechanisms of insect avirulence effectors have remained unknown. Here we identify an insect salivary protein perceived by a plant immune receptor. The BPH14-interacting salivary protein (BISP) from the brown planthopper (Nilaparvata lugens Stål) is secreted into rice (Oryza sativa) during feeding. In susceptible plants, BISP targets O. satvia RLCK185 (OsRLCK185; hereafter Os is used to denote O. satvia-related proteins or genes) to suppress basal defences. In resistant plants, the nucleotide-binding leucine-rich repeat receptor BPH14 directly binds BISP to activate HPR. Constitutive activation of Bph14-mediated immunity is detrimental to plant growth and productivity. The fine-tuning of Bph14-mediated HPR is achieved through direct binding of BISP and BPH14 to the selective autophagy cargo receptor OsNBR1, which delivers BISP to OsATG8 for degradation. Autophagy therefore controls BISP levels. In Bph14 plants, autophagy restores cellular homeostasis by downregulating HPR when feeding by brown planthoppers ceases. We identify an insect saliva protein sensed by a plant immune receptor and discover a three-way interaction system that offers opportunities for developing high-yield, insect-resistant crops.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Kaloshian I, Walling L. Hemipteran and dipteran pests: effectors and plant host immune regulators. J. Integr. Plant Biol. 2016;58:350–361. - PubMed
-
- Hatchett JH, Gallun RL. Genetics of the ability of the Hessian fly, Mayetiola destructor, to survive on wheats having different genes for resistance. Ann. Entomol. Soc. Am. 1970;63:1400–1407.
-
- Oerke EC. Crop losses to pests. J. Agric. Sci. 2006;144:31–43.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

