The Skin Microbiome: Current Techniques, Challenges, and Future Directions
- PMID: 37317196
- PMCID: PMC10223452
- DOI: 10.3390/microorganisms11051222
The Skin Microbiome: Current Techniques, Challenges, and Future Directions
Abstract
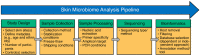
Skin acts as a barrier that promotes the colonization of bacteria, fungi, archaea, and viruses whose membership and function may differ depending on the various specialized niches or micro-environments of the skin. The group of microorganisms inhabiting the skin, also known as the skin microbiome, offers protection against pathogens while actively interacting with the host's immune system. Some members of the skin microbiome can also act as opportunistic pathogens. The skin microbiome is influenced by factors such as skin site, birth mode, genetics, environment, skin products, and skin conditions. The association(s) of the skin microbiome with health and disease has (have) been identified and characterized via culture-dependent and culture-independent methods. Culture-independent methods (such as high-throughput sequencing), in particular, have expanded our understanding of the skin microbiome's role in maintaining health or promoting disease. However, the intrinsic challenges associated with the low microbial biomass and high host content of skin microbiome samples have hindered advancements in the field. In addition, the limitations of current collection and extraction methods and biases derived from sample preparation and analysis have significantly influenced the results and conclusions of many skin microbiome studies. Therefore, the present review discusses the technical challenges associated with the collection and processing of skin microbiome samples, the advantages and disadvantages of current sequencing approaches, and potential future areas of focus for the field.
Keywords: culture-independent methods; metatranscriptomics; shotgun metagenomics; skin microbiome.
Conflict of interest statement
T.M.S.-R. and J.M.M. are current employees of Diversigen, and E.D. and B.L.F. are current employees of DNA Genotek. T.M.S-R., E.B.H. and E.D. are shareholders in OraSure Technologies, the parent company of Diversigen and DNA Genotek.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

