Whole-Genome Sequencing of Shiga Toxin-Producing Escherichia coli for Characterization and Outbreak Investigation
- PMID: 37317272
- PMCID: PMC10224053
- DOI: 10.3390/microorganisms11051298
Whole-Genome Sequencing of Shiga Toxin-Producing Escherichia coli for Characterization and Outbreak Investigation
Abstract
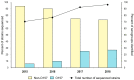
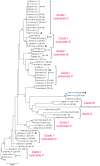
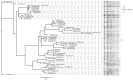
Shiga toxin-producing Escherichia coli (STEC) causes high frequencies of foodborne infections worldwide and has been linked to numerous outbreaks each year. Pulsed-field gel electrophoresis (PFGE) has been the gold standard for surveillance until the recent transition to whole-genome sequencing (WGS). To further understand the genetic diversity and relatedness of outbreak isolates, a retrospective analysis of 510 clinical STEC isolates was conducted. Among the 34 STEC serogroups represented, most (59.6%) belonged to the predominant six non-O157 serogroups. Core genome single nucleotide polymorphism (SNP) analysis differentiated clusters of isolates with similar PFGE patterns and multilocus sequence types (STs). One serogroup O26 outbreak strain and another non-typeable (NT) strain, for instance, were identical by PFGE and clustered together by MLST; however, both were distantly related in the SNP analysis. In contrast, six outbreak-associated serogroup O5 strains clustered with five ST-175 serogroup O5 isolates, which were not part of the same outbreak as determined by PFGE. The use of high-quality SNP analyses enhanced the discrimination of these O5 outbreak strains into a single cluster. In all, this study demonstrates how public health laboratories can more rapidly use WGS and phylogenetics to identify related strains during outbreak investigations while simultaneously uncovering important genetic attributes that can inform treatment practices.
Keywords: Escherichia coli; Shiga toxin; genome sequencing; outbreak; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gould L.H., Mody R.K., Ong K.L., Clogher P., Cronquist A.B., Garman K.N., Lathrop S., Medus C., Spina N.L., Webb T.H., et al. Increased Recognition of Non-O157 Shiga Toxin-producing Escherichia coli Infections in the United States during 2000–2010: Epidemiologic Features and Comparison with E. coli O157 Infections. Foodborne Pathog. Dis. 2013;10:453–460. doi: 10.1089/fpd.2012.1401. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

