16S-23S rRNA Internal Transcribed Spacer Region (ITS) Sequencing: A Potential Molecular Diagnostic Tool for Differentiating Lactococcus garvieae and Lactococcus petauri
- PMID: 37317294
- PMCID: PMC10223780
- DOI: 10.3390/microorganisms11051320
16S-23S rRNA Internal Transcribed Spacer Region (ITS) Sequencing: A Potential Molecular Diagnostic Tool for Differentiating Lactococcus garvieae and Lactococcus petauri
Abstract
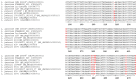
Lactococcus garvieae is the etiological agent of lactococcosis, a clinically and economically significant infectious disease affecting farmed rainbow trout. L. garvieae had been considered the only cause of lactococcosis for a long time; however, L. petauri, another species of the genus Lactococcus, has lately been linked to the same disease. The genomes and biochemical profiles of L. petauri and L. garvieae have a high degree of similarity. Traditional diagnostic tests currently available cannot distinguish between these two species. The aim of this study was to use the transcribed spacer (ITS) region between 16S rRNA and 23S rRNA as a potential useful molecular target to differentiate L. garvieae from L. petauri, saving time and money compared to genomics methods currently used as diagnostic tools for accurate discrimination between these two species. The ITS region of 82 strains was amplified and sequenced. The amplified fragments varied in size from 500 to 550 bp. Based on the sequence, seven SNPs were identified that separate L. garvieae from L. petauri. The 16S-23S rRNA ITS region has enough resolution to distinguish between closely related L. garvieae and L. petauri and it can be used as a diagnostic marker to quickly identify the pathogens in a lactococcosis outbreak.
Keywords: 16S-23S internal transcribed spacer region; Lactococcus garvieae; Lactococcus petauri; diagnostic technique; genome; lactococcosis.
Conflict of interest statement
The authors declare no conflict of interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Castro R., Reguera-Brito M., López-Campos G.H., Blanco M.M., Aguado-Urda M., Fernández-Garayzábal J.F., Gibello A. How does temperature influence the development of lactococcosis? Transcriptomic and immunoproteomic in vitro approaches. J. Fish Dis. 2017;40:1285–1297. doi: 10.1111/jfd.12601. - DOI - PubMed
-
- Goodman L.B., Lawton M.R., Franklin-Guild R.J., Anderson R.R., Schaan L., Thachil A.J., Wiedmann M., Miller C.B., Alcaine S.D., Kovac J. Lactococcus petauri sp. nov., isolated from an abscess of a sugar glider. Int. J. Syst. Evol. Microbiol. 2017;67:4397–4404. doi: 10.1099/ijsem.0.002303. - DOI - PMC - PubMed
-
- Savvidis K., Anatoliotis C., Kanaki Z., Vafeas G. Epizootic outbreaks of lactococcosis disease in rainbow trout, Oncorhynchus mykiss (Walbaum), culture in Greece. Bull. Eur. Assoc. Fish Pathol. 2007;27:223–228.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

