Characterization of genetic variants in the EGLN1/PHD2 gene identified in a European collection of patients with erythrocytosis
- PMID: 37317877
- PMCID: PMC10620589
- DOI: 10.3324/haematol.2023.282913
Characterization of genetic variants in the EGLN1/PHD2 gene identified in a European collection of patients with erythrocytosis
Abstract
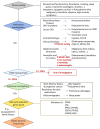
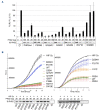
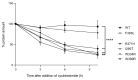
Hereditary erythrocytosis is a rare hematologic disorder characterized by an excess of red blood cell production. Here we describe a European collaborative study involving a collection of 2,160 patients with erythrocytosis sequenced in ten different laboratories. We focused our study on the EGLN1 gene and identified 39 germline missense variants including one gene deletion in 47 probands. EGLN1 encodes the PHD2 prolyl 4-hydroxylase, a major inhibitor of hypoxia-inducible factor. We performed a comprehensive study to evaluate the causal role of the identified PHD2 variants: (i) in silico studies of localization, conservation, and deleterious effects; (ii) analysis of hematologic parameters of carriers identified in the UK Biobank; (iii) functional studies of the protein activity and stability; and (iv) a comprehensive study of PHD2 splicing. Altogether, these studies allowed the classification of 16 pathogenic or likely pathogenic mutants in a total of 48 patients and relatives. The in silico studies extended to the variants described in the literature showed that a minority of PHD2 variants can be classified as pathogenic (36/96), without any differences from the variants of unknown significance regarding the severity of the developed disease (hematologic parameters and complications). Here, we demonstrated the great value of federating laboratories working on such rare disorders in order to implement the criteria required for genetic classification, a strategy that should be extended to all hereditary hematologic diseases.
Figures


References
-
- Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464-468. - PubMed
-
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468-472. - PubMed
-
- Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43-54. - PubMed
-
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005(306):re12. - PubMed
-
- D’Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia upregulates prolyl hydroxylase activity: a feedback mechanism that limits HIF-1 responses during reoxygenation. J Biol Chem. 2003;278(40):38183-38187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

