Brain microglia serve as a persistent HIV reservoir despite durable antiretroviral therapy
- PMID: 37317962
- PMCID: PMC10266791
- DOI: 10.1172/JCI167417
Brain microglia serve as a persistent HIV reservoir despite durable antiretroviral therapy
Abstract
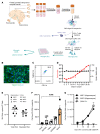
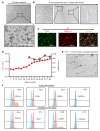
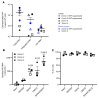
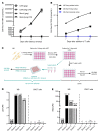
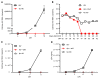
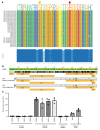
Brain microglia (MG) may serve as a human immunodeficiency virus 1 (HIV) reservoir and ignite rebound viremia following cessation of antiretroviral therapy (ART), but they have yet to be proven to harbor replication-competent HIV. Here, we isolated brain myeloid cells (BrMCs) from nonhuman primates and rapid autopsy of people with HIV (PWH) on ART and sought evidence of persistent viral infection. BrMCs predominantly displayed microglial markers, in which up to 99.9% of the BrMCs were TMEM119+ MG. Total and integrated SIV or HIV DNA was detectable in the MG, with low levels of cell-associated viral RNA. Provirus in MG was highly sensitive to epigenetic inhibition. Outgrowth virus from parietal cortex MG in an individual with HIV productively infected both MG and PBMCs. This inducible, replication-competent virus and virus from basal ganglia proviral DNA were closely related but highly divergent from variants in peripheral compartments. Phenotyping studies characterized brain-derived virus as macrophage tropic based on the ability of the virus to infect cells expressing low levels of CD4. The lack of genetic diversity in virus from the brain suggests that this macrophage-tropic lineage quickly colonized brain regions. These data demonstrate that MG harbor replication-competent HIV and serve as a persistent reservoir in the brain.
Keywords: AIDS/HIV; Drug therapy.
Conflict of interest statement
Figures


References
-
- Joseph SB, et al. Human immunodeficiency virus type 1 RNA detected in the central nervous system (CNS) after years of suppressive antiretroviral therapy can originate from a replicating CNS reservoir or clonally expanded cells. Clin Infect Dis. 2019;69(8):1345–1352. doi: 10.1093/cid/ciy1066. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- P30 AI050409/AI/NIAID NIH HHS/United States
- UM1 AI164559/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- R21 MH128034/MH/NIMH NIH HHS/United States
- DP2 DA051915/DA/NIDA NIH HHS/United States
- P01 AI169609/AI/NIAID NIH HHS/United States
- UM1 AI164567/AI/NIAID NIH HHS/United States
- P30 MH062512/MH/NIMH NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- R01 DA055491/DA/NIDA NIH HHS/United States
- U42 OD011023/OD/NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- R56 AI158293/AI/NIAID NIH HHS/United States
- U24 MH100928/MH/NIMH NIH HHS/United States
- R01 AI147821/AI/NIAID NIH HHS/United States
- UL1 TR001442/TR/NCATS NIH HHS/United States
- P30 AI036219/AI/NIAID NIH HHS/United States
- P01 AI131385/AI/NIAID NIH HHS/United States
- U01 HL146194/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

