Sticky, Adaptable, and Many-sided: SAM protein versatility in normal and pathological hematopoietic states
- PMID: 37318311
- PMCID: PMC10527593
- DOI: 10.1002/bies.202300022
Sticky, Adaptable, and Many-sided: SAM protein versatility in normal and pathological hematopoietic states
Abstract
With decades of research seeking to generalize sterile alpha motif (SAM) biology, many outstanding questions remain regarding this multi-tool protein module. Recent data from structural and molecular/cell biology has begun to reveal new SAM modes of action in cell signaling cascades and biomolecular condensation. SAM-dependent mechanisms underlie blood-related (hematologic) diseases, including myelodysplastic syndromes and leukemias, prompting our focus on hematopoiesis for this review. With the increasing coverage of SAM-dependent interactomes, a hypothesis emerges that SAM interaction partners and binding affinities work to fine tune cell signaling cascades in developmental and disease contexts, including hematopoiesis and hematologic disease. This review discusses what is known and remains unknown about the standard mechanisms and neoplastic properties of SAM domains and what the future might hold for developing SAM-targeted therapies.
Keywords: cell biology; hematopoiesis; signal transduction; sterile alpha motif; transcription.
© 2023 The Authors. BioEssays published by Wiley Periodicals LLC.
Figures

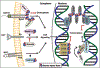
Similar articles
-
Sam Domains in Multiple Diseases.Curr Med Chem. 2020;27(3):450-476. doi: 10.2174/0929867325666181009114445. Curr Med Chem. 2020. PMID: 30306850 Review.
-
Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.Elife. 2018 May 11;7:e35677. doi: 10.7554/eLife.35677. Elife. 2018. PMID: 29749928 Free PMC article.
-
Sterile α-motif domain requirement for cellular signaling and survival.J Biol Chem. 2020 May 15;295(20):7113-7125. doi: 10.1074/jbc.RA119.011895. Epub 2020 Apr 2. J Biol Chem. 2020. PMID: 32241909 Free PMC article.
-
DLC1 SAM domain-binding peptides inhibit cancer cell growth and migration by inactivating RhoA.J Biol Chem. 2020 Jan 10;295(2):645-656. doi: 10.1074/jbc.RA119.011929. Epub 2019 Dec 5. J Biol Chem. 2020. PMID: 31806702 Free PMC article.
-
Plant SAM-Domain Proteins Start to Reveal Their Roles.Trends Plant Sci. 2017 Aug;22(8):718-725. doi: 10.1016/j.tplants.2017.06.006. Epub 2017 Jun 28. Trends Plant Sci. 2017. PMID: 28668510 Review.
Cited by
-
The SAMD1 transcription factor coordinates hematopoietic lineage differentiation and H3K4 methylation status.Blood Adv. 2025 Aug 12;9(15):3988-4003. doi: 10.1182/bloodadvances.2024015627. Blood Adv. 2025. PMID: 40402130 Free PMC article.
-
Cooperation of a polymerizing SAM domain and an intrinsically disordered region enables full SAMD1 function on chromatin.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf259. doi: 10.1093/nar/gkaf259. Nucleic Acids Res. 2025. PMID: 40183636 Free PMC article.
-
Signaling mechanisms and cis -regulatory control of Samd14 in erythroid regeneration.Curr Opin Hematol. 2025 Jul 1;32(4):206-212. doi: 10.1097/MOH.0000000000000873. Epub 2025 Apr 24. Curr Opin Hematol. 2025. PMID: 40293138 Review.
References
-
- DeCamillis M, et al., The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev, 1992. 6(2): p. 223–32. - PubMed
-
- Mackereth CD, et al., Diversity in Structure and Function of the Ets Family PNT Domains. Journal of Molecular Biology, 2004. 342(4): p. 1249–1264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

