Sticky, Adaptable, and Many-sided: SAM protein versatility in normal and pathological hematopoietic states
- PMID: 37318311
- PMCID: PMC10527593
- DOI: 10.1002/bies.202300022
Sticky, Adaptable, and Many-sided: SAM protein versatility in normal and pathological hematopoietic states
Abstract
With decades of research seeking to generalize sterile alpha motif (SAM) biology, many outstanding questions remain regarding this multi-tool protein module. Recent data from structural and molecular/cell biology has begun to reveal new SAM modes of action in cell signaling cascades and biomolecular condensation. SAM-dependent mechanisms underlie blood-related (hematologic) diseases, including myelodysplastic syndromes and leukemias, prompting our focus on hematopoiesis for this review. With the increasing coverage of SAM-dependent interactomes, a hypothesis emerges that SAM interaction partners and binding affinities work to fine tune cell signaling cascades in developmental and disease contexts, including hematopoiesis and hematologic disease. This review discusses what is known and remains unknown about the standard mechanisms and neoplastic properties of SAM domains and what the future might hold for developing SAM-targeted therapies.
Keywords: cell biology; hematopoiesis; signal transduction; sterile alpha motif; transcription.
© 2023 The Authors. BioEssays published by Wiley Periodicals LLC.
Figures

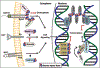
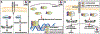
References
-
- DeCamillis M, et al., The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev, 1992. 6(2): p. 223–32. - PubMed
-
- Mackereth CD, et al., Diversity in Structure and Function of the Ets Family PNT Domains. Journal of Molecular Biology, 2004. 342(4): p. 1249–1264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

