Development and Characterization of Potent Succinate Receptor Fluorescent Tracers
- PMID: 37318348
- PMCID: PMC10350927
- DOI: 10.1021/acs.jmedchem.3c00552
Development and Characterization of Potent Succinate Receptor Fluorescent Tracers
Abstract
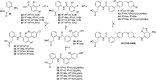
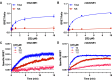
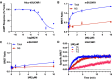
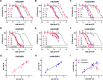
The succinate receptor (SUCNR1) has emerged as a potential target for the treatment of various metabolic and inflammatory diseases, including hypertension, inflammatory bowel disease, and rheumatoid arthritis. While several ligands for this receptor have been reported, species differences in pharmacology between human and rodent orthologs have limited the validation of SUCNR1's therapeutic potential. Here, we describe the development of the first potent fluorescent tool compounds for SUCNR1 and use these to define key differences in ligand binding to human and mouse SUCNR1. Starting from known agonist scaffolds, we developed a potent agonist tracer, TUG-2384 (22), with affinity for both human and mouse SUCNR1. In addition, we developed a novel antagonist tracer, TUG-2465 (46), which displayed high affinity for human SUCNR1. Using 46 we demonstrate that three humanizing mutations on mouse SUCNR1, N181.31E, K2697.32N, and G84EL1W, are sufficient to restore high-affinity binding of SUCNR1 antagonists to the mouse receptor ortholog.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Rexen Ulven E.; Trauelsen M.; Brvar M.; Luckmann M.; Bielefeldt L. O.; Jensen L. K. I.; Schwartz T. W.; Frimurer T. M. Structure-Activity Investigations and Optimisations of Non-metabolite Agonists for the Succinate Receptor 1. Sci. Rep. 2018, 8, 10010.10.1038/s41598-018-28263-7. - DOI - PMC - PubMed
-
- Gnana-Prakasam J. P.; Ananth S.; Prasad P. D.; Zhang M.; Atherton S. S.; Martin P. M.; Smith S. B.; Ganapathy V. Expression and iron-dependent regulation of succinate receptor GPR91 in retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3751–3758. 10.1167/iovs.10-6722. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information

