A novel anoikis-related risk model predicts prognosis in patients with colorectal cancer and responses to different immunotherapy strategies
- PMID: 37318595
- PMCID: PMC11796863
- DOI: 10.1007/s00432-023-04945-2
A novel anoikis-related risk model predicts prognosis in patients with colorectal cancer and responses to different immunotherapy strategies
Abstract
Purpose: We aimed to study the role of anoikis-related genes (ARGs) in colorectal cancer (CRC) using bioinformatics.
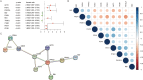
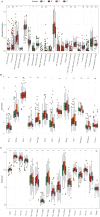
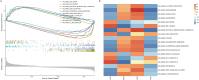
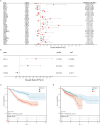
Methods: GSE39582 and GSE39084, which collectively contain 363 CRC samples, were downloaded from the NCBI Gene Expression Omnibus (GEO) database as a test set. TCGA-COADREAD, with 376 CRC samples, was downloaded from the UCSC database as a validation set. Univariate Cox regression analysis was used to screen for ARGs that were significantly associated with prognosis. The top 10 ARGs were used to classify the samples into different subtypes based on unsupervised cluster analysis. The immune environments of the different subtypes were analyzed. ARGs that were significantly associated with CRC prognosis were used to construct a risk model. Univariate and multivariate Cox regression analyses were used to screen independent prognostic factors and construct a nomogram.
Results: Four anoikis-related subtypes (ARSs) with differential prognoses and immune microenvironments were identified. KRAS and epithelial-mesenchymal transition pathways were enriched in subtype B, which had the worst prognosis. Three ARGs (DLG1, AKT3, and LPAR1) were used to construct the risk model. Both the test and validation sets showed worse outcomes for patients in the high-risk group than those in the low-risk group. Risk score was found to be an independent prognostic factor for CRC. Moreover, there was a difference in drug sensitivity between the high- and low-risk groups.
Conclusion: The identified ARGs and risk scores were associated with CRC prognosis and could predict the responses of patients with CRC to immunotherapy strategies.
Keywords: Anoikis; Colorectal cancer; Epithelial-mesenchymal transition; Immune microenvironment; Risk model.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



Similar articles
-
Characterization of novel anoikis-related genes as prognostic biomarkers and key determinants of the immune microenvironment in esophageal cancer.Front Immunol. 2025 Jul 11;16:1599171. doi: 10.3389/fimmu.2025.1599171. eCollection 2025. Front Immunol. 2025. PMID: 40746552 Free PMC article.
-
Identification and verification of a novel anoikis-related gene signature with prognostic significance in clear cell renal cell carcinoma.J Cancer Res Clin Oncol. 2023 Oct;149(13):11661-11678. doi: 10.1007/s00432-023-05012-6. Epub 2023 Jul 5. J Cancer Res Clin Oncol. 2023. PMID: 37402968 Free PMC article.
-
SLC4A4 Moulds the Inflammatory Tumor Microenvironment and Predicts Therapeutic Expectations in Colorectal Cancer.Curr Med Chem. 2025;32(19):3861-3878. doi: 10.2174/0109298673277357231218070812. Curr Med Chem. 2025. PMID: 38310390
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
Cited by
-
TJP3 promotes T cell immunity escape and chemoresistance in breast cancer: a comprehensive analysis of anoikis-based prognosis prediction and drug sensitivity stratification.Aging (Albany NY). 2023 Nov 10;15(22):12890-12906. doi: 10.18632/aging.205208. Epub 2023 Nov 10. Aging (Albany NY). 2023. PMID: 37950731 Free PMC article.
-
A novel anoikis related gene prognostic model for colorectal cancer based on single cell sequencing and bulk transcriptome analyses.Sci Rep. 2025 Aug 18;15(1):30155. doi: 10.1038/s41598-025-15389-8. Sci Rep. 2025. PMID: 40820175 Free PMC article.
-
Anoikis regulator GLI2 promotes NC cell immunity escape by TGF-β-mediated non-classic hedgehog signaling in colorectal cancer: based on artificial intelligence and big data analysis.Aging (Albany NY). 2023 Dec 29;15(24):14733-14748. doi: 10.18632/aging.205283. Epub 2023 Dec 29. Aging (Albany NY). 2023. PMID: 38159250 Free PMC article.
-
Multi-cohort validation of Ascore: an anoikis-based prognostic signature for predicting disease progression and immunotherapy response in bladder cancer.Mol Cancer. 2024 Feb 10;23(1):30. doi: 10.1186/s12943-024-01945-9. Mol Cancer. 2024. PMID: 38341586 Free PMC article.
-
Identification of the molecular subtypes and signatures to predict the prognosis, biological functions, and therapeutic response based on the anoikis-related genes in colorectal cancer.Cancer Med. 2024 May;13(10):e7315. doi: 10.1002/cam4.7315. Cancer Med. 2024. PMID: 38785271 Free PMC article.
References
-
- Alexander PG, McMillan DC, Park JH (2021) A meta-analysis of CD274 (PD-L1) assessment and prognosis in colorectal cancer and its role in predicting response to anti-PD-1 therapy. Crit Rev Oncol/hematol 157:103147. 10.1016/j.critrevonc.2020.103147 - PubMed
-
- Allemani C, Matsuda T, Di Carlo V et al (2018) Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet (london, England) 391:1023–1075. 10.1016/s0140-6736(17)33326-3 - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

