Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages - United States, January 2022-May 2023
- PMID: 37319011
- PMCID: PMC10328465
- DOI: 10.15585/mmwr.mm7224a2
Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages - United States, January 2022-May 2023
Abstract
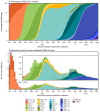
CDC has used national genomic surveillance since December 2020 to monitor SARS-CoV-2 variants that have emerged throughout the COVID-19 pandemic, including the Omicron variant. This report summarizes U.S. trends in variant proportions from national genomic surveillance during January 2022-May 2023. During this period, the Omicron variant remained predominant, with various descendant lineages reaching national predominance (>50% prevalence). During the first half of 2022, BA.1.1 reached predominance by the week ending January 8, 2022, followed by BA.2 (March 26), BA.2.12.1 (May 14), and BA.5 (July 2); the predominance of each variant coincided with surges in COVID-19 cases. The latter half of 2022 was characterized by the circulation of sublineages of BA.2, BA.4, and BA.5 (e.g., BQ.1 and BQ.1.1), some of which independently acquired similar spike protein substitutions associated with immune evasion. By the end of January 2023, XBB.1.5 became predominant. As of May 13, 2023, the most common circulating lineages were XBB.1.5 (61.5%), XBB.1.9.1 (10.0%), and XBB.1.16 (9.4%); XBB.1.16 and XBB.1.16.1 (2.4%), containing the K478R substitution, and XBB.2.3 (3.2%), containing the P521S substitution, had the fastest doubling times at that point. Analytic methods for estimating variant proportions have been updated as the availability of sequencing specimens has declined. The continued evolution of Omicron lineages highlights the importance of genomic surveillance to monitor emerging variants and help guide vaccine development and use of therapeutics.
Conflict of interest statement
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
Figures
References
-
- Lambrou AS, Shirk P, Steele MK, et al.; Strain Surveillance and Emerging Variants Bioinformatic Working Group; Strain Surveillance and Emerging Variants NS3 Working Group. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants—United States, June 2021–January 2022. MMWR Morb Mortal Wkly Rep 2022;71:206–11. 10.15585/mmwr.mm7106a4 - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

