The non-cell-autonomous function of ID1 promotes AML progression via ANGPTL7 from the microenvironment
- PMID: 37319434
- PMCID: PMC10644073
- DOI: 10.1182/blood.2022019537
The non-cell-autonomous function of ID1 promotes AML progression via ANGPTL7 from the microenvironment
Abstract
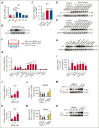
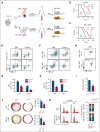
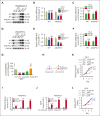
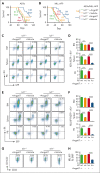
The bone marrow microenvironment (BMM) can regulate leukemia stem cells (LSCs) via secreted factors. Increasing evidence suggests that dissecting the mechanisms by which the BMM maintains LSCs may lead to the development of effective therapies for the eradication of leukemia. Inhibitor of DNA binding 1 (ID1), a key transcriptional regulator in LSCs, previously identified by us, controls cytokine production in the BMM, but the role of ID1 in acute myeloid leukemia (AML) BMM remains obscure. Here, we report that ID1 is highly expressed in the BMM of patients with AML, especially in BM mesenchymal stem cells, and that the high expression of ID1 in the AML BMM is induced by BMP6, secreted from AML cells. Knocking out ID1 in mesenchymal cells significantly suppresses the proliferation of cocultured AML cells. Loss of Id1 in the BMM results in impaired AML progression in AML mouse models. Mechanistically, we found that Id1 deficiency significantly reduces SP1 protein levels in mesenchymal cells cocultured with AML cells. Using ID1-interactome analysis, we found that ID1 interacts with RNF4, an E3 ubiquitin ligase, and causes a decrease in SP1 ubiquitination. Disrupting the ID1-RNF4 interaction via truncation in mesenchymal cells significantly reduces SP1 protein levels and delays AML cell proliferation. We identify that the target of Sp1, Angptl7, is the primary differentially expression protein factor in Id1-deficient BM supernatant fluid to regulate AML progression in mice. Our study highlights the critical role of ID1 in the AML BMM and aids the development of therapeutic strategies for AML.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

