Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection
- PMID: 37322000
- PMCID: PMC10267556
- DOI: 10.1038/s41467-023-39267-x
Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection
Abstract
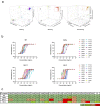
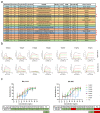
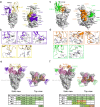
The SARS-CoV-2 Omicron variant evades most currently approved neutralizing antibodies (nAbs) and caused drastic decrease of plasma neutralizing activity elicited by vaccination or prior infection, urging the need for the development of pan-variant antivirals. Breakthrough infection induces a hybrid immunological response with potentially broad, potent and durable protection against variants, therefore, convalescent plasma from breakthrough infection may provide a broadened repertoire for identifying elite nAbs. We performed single-cell RNA sequencing (scRNA-seq) and BCR sequencing (scBCR-seq) of B cells from BA.1 breakthrough-infected patients who received 2 or 3 previous doses of inactivated vaccine. Elite nAbs, mainly derived from the IGHV2-5 and IGHV3-66/53 germlines, showed potent neutralizing activity across Wuhan-Hu-1, Delta, Omicron sublineages BA.1 and BA.2 at picomolar NT50 values. Cryo-EM analysis revealed diverse modes of spike recognition and guides the design of cocktail therapy. A single injection of paired antibodies cocktail provided potent protection in the K18-hACE2 transgenic female mouse model of SARS-CoV-2 infection.
© 2023. The Author(s).
Conflict of interest statement
T.C., Z.R., Y. Guo., P.Z., D.X., X. Chen., X.W., Y.L., X. Cheng., G.Z., X.X., F.L., Q.Y., Z.L., J.T., and J.L. declare the following competing interests: patent has been filed for some of the antibodies presented here (patent application number: 202211015148.3). All other authors declare no competing interests.
Figures

Similar articles
-
Significant neutralizing escapes of Omicron and its sublineages in SARS-CoV-2-infected individuals vaccinated with inactivated vaccines.J Med Virol. 2023 Feb;95(2):e28516. doi: 10.1002/jmv.28516. J Med Virol. 2023. PMID: 36680413
-
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone.Int J Mol Sci. 2022 Jul 12;23(14):7675. doi: 10.3390/ijms23147675. Int J Mol Sci. 2022. PMID: 35887023 Free PMC article.
-
Distinct features of SARS-CoV-2 humoral immunity against Omicron breakthrough infection.Vaccine. 2023 Nov 13;41(47):7019-7025. doi: 10.1016/j.vaccine.2023.10.035. Epub 2023 Oct 17. Vaccine. 2023. PMID: 37858449
-
Omicron BA.1 breakthrough infections in inactivated COVID-19 vaccine recipients induced distinct pattern of antibody and T cell responses to different Omicron sublineages.Emerg Microbes Infect. 2023 Dec;12(1):2202263. doi: 10.1080/22221751.2023.2202263. Emerg Microbes Infect. 2023. PMID: 37037791 Free PMC article.
-
Exposure to BA.4/5 S protein drives neutralization of Omicron BA.1, BA.2, BA.2.12.1, and BA.4/5 in vaccine-experienced humans and mice.Sci Immunol. 2022 Dec 23;7(78):eade9888. doi: 10.1126/sciimmunol.ade9888. Epub 2022 Dec 23. Sci Immunol. 2022. PMID: 36378074 Free PMC article.
Cited by
-
Structural Immunology of SARS-CoV-2.Immunol Rev. 2025 Jan;329(1):e13431. doi: 10.1111/imr.13431. Epub 2024 Dec 27. Immunol Rev. 2025. PMID: 39731211 Free PMC article. Review.
-
Single-cell transcriptome atlas of peripheral immune features to Omicron breakthrough infection under booster vaccination strategies.Front Immunol. 2025 Jan 6;15:1460442. doi: 10.3389/fimmu.2024.1460442. eCollection 2024. Front Immunol. 2025. PMID: 39835127 Free PMC article.
-
Molecular mechanism and structure-guided humanization of a broadly neutralizing antibody against SFTSV.PLoS Pathog. 2024 Sep 25;20(9):e1012550. doi: 10.1371/journal.ppat.1012550. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39321193 Free PMC article.
-
The crystal structure of coronavirus RBD-TMPRSS2 complex provides basis for the discovery of therapeutic antibodies.Nat Commun. 2025 Jul 18;16(1):6636. doi: 10.1038/s41467-025-62023-2. Nat Commun. 2025. PMID: 40681508 Free PMC article.
-
Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering.Proc Natl Acad Sci U S A. 2025 Feb 11;122(6):e2406659122. doi: 10.1073/pnas.2406659122. Epub 2025 Feb 5. Proc Natl Acad Sci U S A. 2025. PMID: 39908098 Free PMC article.
References
-
- Weekly epidemiological update on COVID-19 - 15 February 2023. https://www.who.int/publications/m/item/weekly-epidemiological-update-on....
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

