ChAdOx1 nCoV-19 (AZD1222) vaccine-induced Fc receptor binding tracks with differential susceptibility to COVID-19
- PMID: 37322179
- PMCID: PMC10307634
- DOI: 10.1038/s41590-023-01513-1
ChAdOx1 nCoV-19 (AZD1222) vaccine-induced Fc receptor binding tracks with differential susceptibility to COVID-19
Abstract
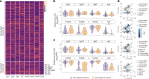
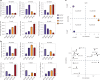
Despite the success of COVID-19 vaccines, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern have emerged that can cause breakthrough infections. Although protection against severe disease has been largely preserved, the immunological mediators of protection in humans remain undefined. We performed a substudy on the ChAdOx1 nCoV-19 (AZD1222) vaccinees enrolled in a South African clinical trial. At peak immunogenicity, before infection, no differences were observed in immunoglobulin (Ig)G1-binding antibody titers; however, the vaccine induced different Fc-receptor-binding antibodies across groups. Vaccinees who resisted COVID-19 exclusively mounted FcγR3B-binding antibodies. In contrast, enhanced IgA and IgG3, linked to enriched FcγR2B binding, was observed in individuals who experienced breakthrough. Antibodies unable to bind to FcγR3B led to immune complex clearance and resulted in inflammatory cascades. Differential antibody binding to FcγR3B was linked to Fc-glycosylation differences in SARS-CoV-2-specific antibodies. These data potentially point to specific FcγR3B-mediated antibody functional profiles as critical markers of immunity against COVID-19.
© 2023. The Author(s).
Conflict of interest statement
G.A. is a founder/equity holder in Seromyx Systems Inc. and Leyden Labs. She has served as a scientific advisor for Sanofi Vaccines and has collaborative agreements with GSK, Merck, Abbvie, Sanofi, Medicago, BioNtech, Moderna, BMS, Novavax, SK Bioscience, Gilead and Sanaria. The remaining authors declare no conflicting interests.
Figures




References
-
- COVID Data Tracker. Centers for Disease Control and Preventionhttps://covid.cdc.gov/covid-data-tracker/#datatracker-home (2021).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

