Molecular and clinical characterization of PTRF in glioma via 1,022 samples
- PMID: 37322408
- PMCID: PMC10273567
- DOI: 10.1186/s12885-023-11001-2
Molecular and clinical characterization of PTRF in glioma via 1,022 samples
Abstract
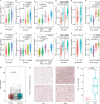
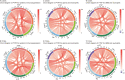
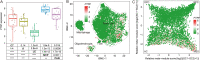
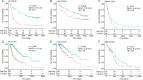
Polymerase I and transcript release factor (PTRF) plays a role in the regulation of gene expression and the release of RNA transcripts during transcription, which have been associated with various human diseases. However, the role of PTRF in glioma remains unclear. In this study, RNA sequencing (RNA-seq) data (n = 1022 cases) and whole-exome sequencing (WES) data (n = 286 cases) were used to characterize the PTRF expression features. Gene ontology (GO) functional enrichment analysis was used to assess the biological implication of changes in PTRF expression. As a result, the expression of PTRF was associated with malignant progression in gliomas. Meanwhile, somatic mutational profiles and copy number variations (CNV) revealed the glioma subtypes classified by PTRF expression showed distinct genomic alteration. Furthermore, GO functional enrichment analysis suggested that PTRF expression was associated with cell migration and angiogenesis, particularly during an immune response. Survival analysis confirmed that a high expression of PTRF is associated with a poor prognosis. In summary, PTRF may be a valuable factor for the diagnosis and treatment target of glioma.
Keywords: Glioma; Immune response; PTRF; Prognosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas.EBioMedicine. 2018 Sep;35:233-243. doi: 10.1016/j.ebiom.2018.08.012. Epub 2018 Aug 18. EBioMedicine. 2018. PMID: 30131308 Free PMC article.
-
PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma.Front Immunol. 2022 Jan 6;12:802795. doi: 10.3389/fimmu.2021.802795. eCollection 2021. Front Immunol. 2022. PMID: 35069587 Free PMC article.
-
Molecular Characterization and Clinical Relevance of ANXA1 in Gliomas via 1,018 Chinese Cohort Patients.Front Cell Dev Biol. 2021 Nov 29;9:777182. doi: 10.3389/fcell.2021.777182. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34912807 Free PMC article.
-
PTRF/cavin-1 remodels phospholipid metabolism to promote tumor proliferation and suppress immune responses in glioblastoma by stabilizing cPLA2.Neuro Oncol. 2021 Mar 25;23(3):387-399. doi: 10.1093/neuonc/noaa255. Neuro Oncol. 2021. PMID: 33140095 Free PMC article.
-
Emerging role of polymerase-1 and transcript release factor (PTRF/ Cavin-1) in health and disease.Cell Tissue Res. 2014 Sep;357(3):505-13. doi: 10.1007/s00441-014-1964-z. Epub 2014 Aug 9. Cell Tissue Res. 2014. PMID: 25107607 Review.
Cited by
-
Using parenclitic networks on phaeochromocytoma and paraganglioma tumours provides novel insights on global DNA methylation.Sci Rep. 2024 Dec 2;14(1):29958. doi: 10.1038/s41598-024-81486-9. Sci Rep. 2024. PMID: 39622952 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

