Assignment of the somatic A/B compartments to chromatin domains in giant transcriptionally active lampbrush chromosomes
- PMID: 37322523
- PMCID: PMC10268536
- DOI: 10.1186/s13072-023-00499-2
Assignment of the somatic A/B compartments to chromatin domains in giant transcriptionally active lampbrush chromosomes
Abstract
Background: The three-dimensional configuration of the eukaryotic genome is an emerging area of research. Chromosome conformation capture outlined genome segregation into large scale A and B compartments corresponding mainly to transcriptionally active and repressive chromatin. It remains unknown how the compartmentalization of the genome changes in growing oocytes of animals with hypertranscriptional type of oogenesis. Such oocytes are characterized by highly elongated chromosomes, called lampbrush chromosomes, which acquire a typical chromomere-loop appearance, representing one of the classical model systems for exploring the structural and functional organization of chromatin domains.
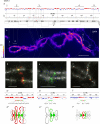
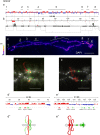
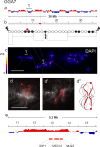
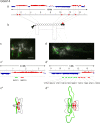
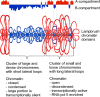
Results: Here, we compared the distribution of A/B compartments in chicken somatic cells with chromatin domains in lampbrush chromosomes. We found that in lampbrush chromosomes, the extended chromatin domains, restricted by compartment boundaries in somatic cells, disintegrate into individual chromomeres. Next, we performed FISH-mapping of the genomic loci, which belong to A or B chromatin compartments as well as to A/B compartment transition regions in embryonic fibroblasts on isolated lampbrush chromosomes. We found, that in chicken lampbrush chromosomes, clusters of dense compact chromomeres bearing short lateral loops and enriched with repressive epigenetic modifications generally correspond to constitutive B compartments in somatic cells. A compartments align with lampbrush chromosome segments with smaller, less compact chromomeres, longer lateral loops, and a higher transcriptional status. Clusters of small loose chromomeres with relatively long lateral loops show no obvious correspondence with either A or B compartment identity. Some genes belonging to facultative B (sub-) compartments can be tissue-specifically transcribed during oogenesis, forming distinct lateral loops.
Conclusions: Here, we established a correspondence between the A/B compartments in somatic interphase nucleus and chromatin segments in giant lampbrush chromosomes from diplotene stage oocytes. The chromomere-loop structure of the genomic regions corresponding to interphase A and B compartments reveals the difference in how they are organized at the level of chromatin domains. The results obtained also suggest that gene-poor regions tend to be packed into chromomeres.
Keywords: A/B compartments; Chicken genome; Chromomere; Chromosome conformation capture; FISH-mapping; Hypertranscription; Lampbrush chromosomes; Meiotic chromosomes; Oocyte nucleus; Transcription loops.
© 2023. The Author(s).
Conflict of interest statement
None of the authors have any competing interests.
Figures

Similar articles
-
Comparison of the somatic TADs and lampbrush chromomere-loop complexes in transcriptionally active prophase I oocytes.Chromosoma. 2022 Dec;131(4):207-223. doi: 10.1007/s00412-022-00780-5. Epub 2022 Aug 29. Chromosoma. 2022. PMID: 36031655
-
New Insights Into Chromomere Organization Provided by Lampbrush Chromosome Microdissection and High-Throughput Sequencing.Front Genet. 2020 Feb 17;11:57. doi: 10.3389/fgene.2020.00057. eCollection 2020. Front Genet. 2020. PMID: 32127797 Free PMC article.
-
Mapping epigenetic modifications on chicken lampbrush chromosomes.Mol Cytogenet. 2020 Aug 3;13:32. doi: 10.1186/s13039-020-00496-0. eCollection 2020. Mol Cytogenet. 2020. PMID: 32774459 Free PMC article.
-
Avian lampbrush chromosomes: a powerful tool for exploration of genome expression.Cytogenet Genome Res. 2009;124(3-4):251-67. doi: 10.1159/000218130. Epub 2009 Jun 25. Cytogenet Genome Res. 2009. PMID: 19556778 Review.
-
[Changes in the relative arrangement of the chromosomes and nucleolus in developing mammalian oocytes during meiotic prophase I in relation to functional changes in the oocytes].Tsitologiia. 1992;34(11-12):3-23. Tsitologiia. 1992. PMID: 1302392 Review. Russian.
Cited by
-
Lampbrush chromosomes of Danio rerio.Chromosome Res. 2025 Jan 16;33(1):2. doi: 10.1007/s10577-024-09761-z. Chromosome Res. 2025. PMID: 39815120
-
Biological functions and clinic significance of SAF‑A (Review).Biomed Rep. 2024 Apr 10;20(6):88. doi: 10.3892/br.2024.1776. eCollection 2024 Jun. Biomed Rep. 2024. PMID: 38665420 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

