Vitamin B12 produced by Cetobacterium somerae improves host resistance against pathogen infection through strengthening the interactions within gut microbiota
- PMID: 37322528
- PMCID: PMC10268390
- DOI: 10.1186/s40168-023-01574-2
Vitamin B12 produced by Cetobacterium somerae improves host resistance against pathogen infection through strengthening the interactions within gut microbiota
Abstract
Background: Pathogen infections seriously affect host health, and the use of antibiotics increases the risk of the emergence of drug-resistant bacteria and also increases environmental and health safety risks. Probiotics have received much attention for their excellent ability to prevent pathogen infections. Particularly, explaining mechanism of action of probiotics against pathogen infections is important for more efficient and rational use of probiotics and the maintenance of host health.
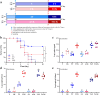
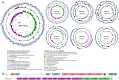
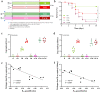
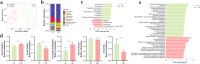
Results: Here, we describe the impacts of probiotic on host resistance to pathogen infections. Our findings revealed that (I) the protective effect of oral supplementation with B. velezensis against Aeromonas hydrophila infection was dependent on gut microbiota, specially the anaerobic indigenous gut microbe Cetobacterium; (II) Cetobacterium was a sensor of health, especially for fish infected with pathogenic bacteria; (III) the genome resolved the ability of Cetobacterium somerae CS2105-BJ to synthesize vitamin B12 de novo, while in vivo and in vitro metabolism assays also showed the ability of Cetobacterium somerae CS2105-BJ to produce vitamin B12; (IV) the addition of vitamin B12 significantly altered the gut redox status and the gut microbiome structure and function, and then improved the stability of the gut microbial ecological network, and enhanced the gut barrier tight junctions to prevent the pathogen infection.
Conclusion: Collectively, this study found that the effect of probiotics in enhancing host resistance to pathogen infections depended on function of B12 produced by an anaerobic indigenous gut microbe, Cetobacterium. Furthermore, as a gut microbial regulator, B12 exhibited the ability to strengthen the interactions within gut microbiota and gut barrier tight junctions, thereby improving host resistance against pathogen infection. Video Abstract.
Keywords: Cetobacterium; Co-occurrence network; Gut microbiome; Pathogen resistance; Probiotics; Vitamin B12.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Exploring the protective role of Bacillus velezensis BV1704-Y in zebrafish health and disease resistance against Aeromonas hydrophila infection.Fish Shellfish Immunol. 2024 Sep;152:109789. doi: 10.1016/j.fsi.2024.109789. Epub 2024 Jul 23. Fish Shellfish Immunol. 2024. PMID: 39053585
-
Vitamin B12-producing Cetobacterium: An important biomarker linked to snake hibernation.Int J Biol Macromol. 2025 May;306(Pt 2):141827. doi: 10.1016/j.ijbiomac.2025.141827. Epub 2025 Mar 7. Int J Biol Macromol. 2025. PMID: 40057061
-
Over supplementation with vitamin B12 alters microbe-host interactions in the gut leading to accelerated Citrobacter rodentium colonization and pathogenesis in mice.Microbiome. 2023 Feb 3;11(1):21. doi: 10.1186/s40168-023-01461-w. Microbiome. 2023. PMID: 36737826 Free PMC article.
-
Application of zebrafish in the study of the gut microbiome.Animal Model Exp Med. 2022 Dec;5(4):323-336. doi: 10.1002/ame2.12227. Epub 2022 Apr 12. Animal Model Exp Med. 2022. PMID: 35415967 Free PMC article. Review.
-
A Review on Gut Remediation of Selected Environmental Contaminants: Possible Roles of Probiotics and Gut Microbiota.Nutrients. 2018 Dec 21;11(1):22. doi: 10.3390/nu11010022. Nutrients. 2018. PMID: 30577661 Free PMC article. Review.
Cited by
-
Effects of solid-state fermentation product of yeast supplementation on liver and intestinal health, and resistance of common carp (Cyprinus carpio) against spring viraemia carp virus.Anim Nutr. 2024 Jun 3;18:408-418. doi: 10.1016/j.aninu.2024.04.017. eCollection 2024 Sep. Anim Nutr. 2024. PMID: 39309973 Free PMC article.
-
Multi-Omics Analysis of Gut Microbiome and Host Metabolism in Different Populations of Chinese Alligators (alligator sinensis) During Various Reintroduction Phases.Ecol Evol. 2025 Apr 9;15(4):e71221. doi: 10.1002/ece3.71221. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40212922 Free PMC article.
-
Rationally designed probiotics prevent shrimp white feces syndrome via the probiotics-gut microbiome-immunity axis.NPJ Biofilms Microbiomes. 2024 Apr 11;10(1):40. doi: 10.1038/s41522-024-00509-5. NPJ Biofilms Microbiomes. 2024. PMID: 38605016 Free PMC article.
-
The carotenoid torularhodin alleviates NAFLD by promoting Akkermanisa muniniphila-mediated adenosylcobalamin metabolism.Nat Commun. 2025 Apr 8;16(1):3338. doi: 10.1038/s41467-025-58500-3. Nat Commun. 2025. PMID: 40199868 Free PMC article.
-
A review on comparative analysis of marine and freshwater fish gut microbiomes: insights into environmental impact on gut microbiota.FEMS Microbiol Ecol. 2025 Jan 7;101(1):fiae169. doi: 10.1093/femsec/fiae169. FEMS Microbiol Ecol. 2025. PMID: 39719366 Free PMC article. Review.
References
-
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–14. doi: 10.1038/nrgastro.2014.66. - DOI - PubMed
-
- Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20(4):515–526. doi: 10.1016/j.chom.2016.09.001. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

