Feedback circuits are numerous in embryonic gene regulatory networks and offer a stabilizing influence on evolution of those networks
- PMID: 37322563
- PMCID: PMC10273620
- DOI: 10.1186/s13227-023-00214-y
Feedback circuits are numerous in embryonic gene regulatory networks and offer a stabilizing influence on evolution of those networks
Abstract
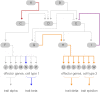
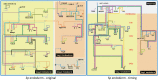
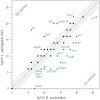
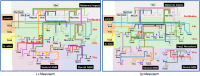
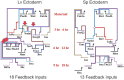
The developmental gene regulatory networks (dGRNs) of two sea urchin species, Lytechinus variegatus (Lv) and Strongylocentrotus purpuratus (Sp), have remained remarkably similar despite about 50 million years since a common ancestor. Hundreds of parallel experimental perturbations of transcription factors with similar outcomes support this conclusion. A recent scRNA-seq analysis suggested that the earliest expression of several genes within the dGRNs differs between Lv and Sp. Here, we present a careful reanalysis of the dGRNs in these two species, paying close attention to timing of first expression. We find that initial expression of genes critical for cell fate specification occurs during several compressed time periods in both species. Previously unrecognized feedback circuits are inferred from the temporally corrected dGRNs. Although many of these feedbacks differ in location within the respective GRNs, the overall number is similar between species. We identify several prominent differences in timing of first expression for key developmental regulatory genes; comparison with a third species indicates that these heterochronies likely originated in an unbiased manner with respect to embryonic cell lineage and evolutionary branch. Together, these results suggest that interactions can evolve even within highly conserved dGRNs and that feedback circuits may buffer the effects of heterochronies in the expression of key regulatory genes.
Keywords: Embryonic specification; Evolutionary mechanism; Feedback circuits; Gene regulatory networks; Sea urchin development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity. Malden: Blackwell; 2001.
-
- Wilkins AS. The evolution of developmental pathways. Sunderland: Sinauer Associates; 2002.
-
- The DE, Genome R. Gene regulatory networks in development and evolution. San Diego: Academic Press/Elsevier; 2006. p. 289.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

