A metagenome-level analysis of a microbial community fermenting ultra-filtered milk permeate
- PMID: 37324413
- PMCID: PMC10263058
- DOI: 10.3389/fbioe.2023.1173656
A metagenome-level analysis of a microbial community fermenting ultra-filtered milk permeate
Abstract
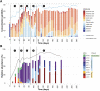
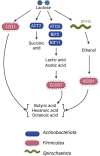
Fermentative microbial communities have the potential to serve as biocatalysts for the conversion of low-value dairy coproducts into renewable chemicals, contributing to a more sustainable global economy. To develop predictive tools for the design and operation of industrially relevant strategies that utilize fermentative microbial communities, there is a need to determine the genomic features of community members that are characteristic to the accumulation of different products. To address this knowledge gap, we performed a 282-day bioreactor experiment with a microbial community that was fed ultra-filtered milk permeate, a low-value coproduct from the dairy industry. The bioreactor was inoculated with a microbial community from an acid-phase digester. A metagenomic analysis was used to assess microbial community dynamics, construct metagenome-assembled genomes (MAGs), and evaluate the potential for lactose utilization and fermentation product synthesis of community members represented by the assembled MAGs. This analysis led us to propose that, in this reactor, members of the Actinobacteriota phylum are important in the degradation of lactose, via the Leloir pathway and the bifid shunt, and the production of acetic, lactic, and succinic acids. In addition, members of the Firmicutes phylum contribute to the chain-elongation-mediated production of butyric, hexanoic, and octanoic acids, with different microbes using either lactose, ethanol, or lactic acid as the growth substrate. We conclude that genes encoding carbohydrate utilization pathways, and genes encoding lactic acid transport into the cell, electron confurcating lactate dehydrogenase, and its associated electron transfer flavoproteins, are genomic features whose presence in Firmicutes needs to be established to infer the growth substrate used for chain elongation.
Keywords: butyric; chain elongation; dairy coproduct; fermentation; hexanoic; lactic; microbiome; succinic.
Copyright © 2023 Walters, Mohan, Myers, Ingle, Donohue and Noguera.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- American Dairy Products Institution (2019). 2018 Dairy products utilization and production trends. Available at: https://www.adpi.org/publications .
-
- American Water Works Association and Water Environment Federation (2005). Standard methods for the examination of water and wastewater. Washington, DC, USA: American Water Works American Public Health Association.
-
- Belenguer A., Duncan S. H., Calder A. G., Holtrop G., Louis P., Lobley G. E., et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72, 3593–3599. 10.1128/AEM.72.5.3593-3599.2006 - DOI - PMC - PubMed
-
- Biddy M. J., Scarlata C., Kinchin C. (2016). Chemicals from biomass: A market assessment of bioproducts with near-term potential. Golden, CO, USA: National Renewable Energy Laboratory. NREL Report NREL/TP-5100-65509.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

