Variation and stability of rhizosphere bacterial communities of Cucumis crops in association with root-knot nematodes infestation
- PMID: 37324672
- PMCID: PMC10266268
- DOI: 10.3389/fpls.2023.1163271
Variation and stability of rhizosphere bacterial communities of Cucumis crops in association with root-knot nematodes infestation
Abstract
Introduction: Root-knot nematodes (RKN) disease is a devastating disease in Cucumis crops production. Existing studies have shown that resistant and susceptible crops are enriched with different rhizosphere microorganisms, and microorganisms enriched in resistant crops can antagonize pathogenic bacteria. However, the characteristics of rhizosphere microbial communities of Cucumis crops after RKN infestation remain largely unknown.
Methods: In this study, we compared the changes in rhizosphere bacterial communities between highly RKN-resistant Cucumis metuliferus (cm3) and highly RKN-susceptible Cucumis sativus (cuc) after RKN infection through a pot experiment.
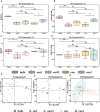
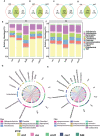
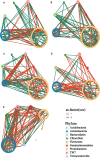
Results: The results showed that the strongest response of rhizosphere bacterial communities of Cucumis crops to RKN infestation occurred during early growth, as evidenced by changes in species diversity and community composition. However, the more stable structure of the rhizosphere bacterial community in cm3 was reflected in less changes in species diversity and community composition after RKN infestation, forming a more complex and positively co-occurrence network than cuc. Moreover, we observed that both cm3 and cuc recruited bacteria after RKN infestation, but the bacteria enriched in cm3 were more abundant including beneficial bacteria Acidobacteria, Nocardioidaceae and Sphingomonadales. In addition, the cuc was enriched with beneficial bacteria Actinobacteria, Bacilli and Cyanobacteria. We also found that more antagonistic bacteria than cuc were screened in cm3 after RKN infestation and most of them were Pseudomonas (Proteobacteria, Pseudomonadaceae), and Proteobacteria were also enriched in cm3 after RKN infestation. We hypothesized that the cooperation between Pseudomonas and the beneficial bacteria in cm3 could inhibit the infestation of RKN.
Discussion: Thus, our results provide valuable insights into the role of rhizosphere bacterial communities on RKN diseases of Cucumis crops, and further studies are needed to clarify the bacterial communities that suppress RKN in Cucumis crops rhizosphere.
Keywords: Cucumis crops; co-occurrence network; rhizosphere bacteria; root-knot nematodes; time dynamic.
Copyright © 2023 Song, Ping, Mao, Zhao, Yang, Li, Xie and Ling.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Rhizosphere Microbiomes from Root Knot Nematode Non-infested Plants Suppress Nematode Infection.Microb Ecol. 2019 Aug;78(2):470-481. doi: 10.1007/s00248-019-01319-5. Epub 2019 Jan 21. Microb Ecol. 2019. PMID: 30666369 Free PMC article.
-
Comprehensive analysis of the WRKY gene family in Cucumis metuliferus and their expression profile in response to an early stage of root knot nematode infection.Front Plant Sci. 2023 Mar 20;14:1143171. doi: 10.3389/fpls.2023.1143171. eCollection 2023. Front Plant Sci. 2023. PMID: 37021316 Free PMC article.
-
Tobacco Root Microbial Community Composition Significantly Associated With Root-Knot Nematode Infections: Dynamic Changes in Microbiota and Growth Stage.Front Microbiol. 2022 Feb 9;13:807057. doi: 10.3389/fmicb.2022.807057. eCollection 2022. Front Microbiol. 2022. PMID: 35222332 Free PMC article.
-
Understanding the dynamic interactions of root-knot nematodes and their host: role of plant growth promoting bacteria and abiotic factors.Front Plant Sci. 2024 Apr 30;15:1377453. doi: 10.3389/fpls.2024.1377453. eCollection 2024. Front Plant Sci. 2024. PMID: 38745927 Free PMC article. Review.
-
Unraveling the enigma of root-knot nematodes: from origins to advanced management strategies in agriculture.Planta. 2024 Jun 26;260(2):36. doi: 10.1007/s00425-024-04464-5. Planta. 2024. PMID: 38922545 Review.
Cited by
-
Rhizosphere microbiome regulation: Unlocking the potential for plant growth.Curr Res Microb Sci. 2024 Nov 22;8:100322. doi: 10.1016/j.crmicr.2024.100322. eCollection 2025. Curr Res Microb Sci. 2024. PMID: 39678067 Free PMC article. Review.
-
Integration of soil microbiology and metabolomics to elucidate the mechanism of the accelerated infestation of tobacco by the root-knot nematode.Front Microbiol. 2024 Aug 23;15:1455880. doi: 10.3389/fmicb.2024.1455880. eCollection 2024. Front Microbiol. 2024. PMID: 39247692 Free PMC article.
References
-
- Aiello D., Restuccia C., Stefani E., Vitale A., Cirvilleri G. (2019). Postharvest biocontrol ability of Pseudomonas synxantha against monilinia fructicola and monilinia fructigena on stone fruit. Postharvest Biol. Technol. 149, 83–89. doi: 10.1016/j.postharvbio.2018.11.020 - DOI
-
- Alberoni D., Gaggìa F., Baffoni L., Modesto M. M., Biavati B., Di Gioia D. (2019). Bifidobacterium xylocopae sp. nov. and bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst. Appl. Microbiol. 42 (2), 205–216. doi: 10.1016/j.syapm.2018.11.005 - DOI - PubMed
-
- Bahareh N., Bouaicha N., Metcalf J. S., Porzani S. J., Konur O. (2021). Plant-cyanobacteria interactions: beneficial and harmful effects of cyanobacterial bioactive compounds on soil-plant systems and subsequent risk to animal and human health. Phytochemistry 192, 112959. doi: 10.1016/j.phytochem.2021.112959 - DOI - PubMed
LinkOut - more resources
Full Text Sources

