An efficient DNAzyme for the fluorescence detection of Vibrio cholerae
- PMID: 37324923
- PMCID: PMC10261802
- DOI: 10.1002/fsn3.3304
An efficient DNAzyme for the fluorescence detection of Vibrio cholerae
Abstract
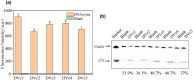
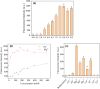
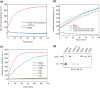
Vibrio cholerae (Vc) causes cholera disease. Vc contamination is widely found in water and aquatic products, and therefore is a serious food safety concern, especially for the seafood industry. In this paper, we attempted the rapid detection of V. cholerae. Nine rounds of in vitro selection using an unmodified DNA library were successfully performed to find specific DNAzymes of Vc. Their activity was evaluated based on a fluorescence assay and gel electrophoresis. Finally, a DNAzyme (named DVc1) with good activity and specificity with a detection limit of 7.2 × 103 CFU/mL of Vc was selected. A simple biosensor was constructed by immobilizing DVc1 and its substrate in shallow circular wells of a 96-well plate using pullulan polysaccharide and trehalose. When the crude extracellular mixture of Vc was added to the detection wells, the fluorescent signal was observed within 20 min. The sensor effectively detected Vc in aquatic products indicating its simplicity and efficiency. This sensitive DNAzyme sensor can be a rapid onsite Vc detection tool.
Keywords: DNAzyme; fluorescence detection; onsite detection; sensitive V. cholerae biosensor.
© 2023 The Authors. Food Science & Nutrition published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Enhanced Detection of Vibrio harveyi Using a Dual-Composite DNAzyme-Based Biosensor.Biosensors (Basel). 2024 Nov 13;14(11):548. doi: 10.3390/bios14110548. Biosensors (Basel). 2024. PMID: 39590007 Free PMC article.
-
Detection of Vibrio vulnificus in Seafood With a DNAzyme-Based Biosensor.Front Microbiol. 2021 Jun 4;12:655845. doi: 10.3389/fmicb.2021.655845. eCollection 2021. Front Microbiol. 2021. PMID: 34149642 Free PMC article.
-
Fast and sensitive graphene oxide-DNAzyme-based biosensor for Vibrio alginolyticus detection.J Fish Dis. 2022 May;45(5):687-697. doi: 10.1111/jfd.13594. Epub 2022 Feb 17. J Fish Dis. 2022. PMID: 35176196
-
Selection of DNAzymes for Sensing Aquatic Bacteria: Vibrio Anguillarum.Anal Chem. 2019 Jun 18;91(12):7887-7893. doi: 10.1021/acs.analchem.9b01707. Epub 2019 May 31. Anal Chem. 2019. PMID: 31117412
-
Rapid and Visualized Detection of Virulence-Related Genes of Vibrio cholerae in Water and Aquatic Products by Loop-Mediated Isothermal Amplification.J Food Prot. 2022 Jan 1;85(1):44-53. doi: 10.4315/JFP-21-182. J Food Prot. 2022. PMID: 34436566
Cited by
-
Aptamer and DNAzyme Based Colorimetric Biosensors for Pathogen Detection.Angew Chem Int Ed Engl. 2025 Jan 21;64(4):e202418725. doi: 10.1002/anie.202418725. Epub 2024 Nov 25. Angew Chem Int Ed Engl. 2025. PMID: 39551709 Free PMC article. Review.
-
DNAzyme hydrogels specifically inhibit the NLRP3 pathway to prevent radiation-induced skin injury in mice.J Nanobiotechnology. 2025 Mar 22;23(1):238. doi: 10.1186/s12951-025-03147-x. J Nanobiotechnology. 2025. PMID: 40119386 Free PMC article.
References
-
- Ali, M. M. , Aguirre, S. D. , Lazim, H. , & Li, Y. (2011). Fluorogenic DNAzyme probes as bacterial indicators. Angewandte Chemie, 123(16), 3835–3838. - PubMed
-
- Ali, M. M. , Wolfe, M. , Tram, K. , Gu, J. , Filipe, C. D. M. , Li, Y. , & Brennan, J. D. (2019). A DNAzyme‐based colorimetric paper sensor for helicobacter pylori. Angewandte Chemie, 131(29), 10012–10016. - PubMed
-
- Ambros, V. , Lee, R. C. , Lavanway, A. , Williams, P. T. , & Jewell, D. (2003). MicroRNAs and other tiny endogenous RNAs in C. elegans. Current Biology, 13(10), 807–818. - PubMed
-
- Balasubramanian, D. , Murcia, S. , Ogbunugafor, C. B. , Gavilan, R. , & Almagro‐Moreno, S. (2021). Cholera dynamics: Lessons from an epidemic. Journal of Medical Microbiology, 70(2), 001298. - PubMed
LinkOut - more resources
Full Text Sources

