Trait profiling and genotype selection in oilseed rape using genotype by trait and genotype by yield*trait approaches
- PMID: 37324925
- PMCID: PMC10261743
- DOI: 10.1002/fsn3.3290
Trait profiling and genotype selection in oilseed rape using genotype by trait and genotype by yield*trait approaches
Abstract
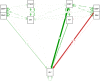
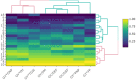
Selection and breeding for high-yielding in oilseed rape have always been one of the leading objectives for oilseed rape breeders. This process becomes more complicated when all quantitative traits are considered in selection in addition to grain yield. In the present study, 18 oilseed rape genotypes along with 2 check cultivars (RGS003 and Dalgan) were evaluated across 16 environments (a combination of 2 years and eight locations) in the tropical climate regions of Iran during 2018-2019 and 2019-2020 cropping seasons. The experiments were conducted in a format of randomized complete block design (RCBD) with three replications. The obtained multienvironmental trial data were utilized to conduct multivariate analysis, genotype by trait (GT) biplot, and genotype by yield*trait (GYT) biplot (Breeding, Genetics and Genomics, 1:2019). The GT and GYT biplot accounted for 55.5% and 93.6% of the total variation in the first two main components. Based on multivariate analysis and GT biplot, pod numbers in plant (PNP) and plant height (PH) were chosen as two key traits in spring oilseed rape genotypes for indirect selection due to high variation, strong positive correlation with grain yield (GY), and their high representatively and discriminability in genotype selection. The mean × stability GT biplot represented G10 (SRL-96-17) as the superior genotype. Based on the mean × stability GYT biplot, eight above-average genotypes were identified that took high scores in stability, high-yielding, and all evaluated quantitative traits at the same time. Based on the superiority index of GYT data, G10 (SRL-96-17) and G5 (SRL-96-11) indicated the best yield-trait combinations profile and ranked above check cultivars and then selected as superior genotypes. Similarly, cluster analysis using the WARD method also separated eight superior genotypes. Based on the result of the present study, GT ad GYT methodologies are recommended for trait profiling and genotype selection in oilseed rape breeding projects, respectively.
Keywords: GT biplot; GYT biplot; high‐yielding; multi environmental trial.
© 2023 The Authors. Food Science & Nutrition published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Amiri Oghan, H. , Sabaghnia, N. , Rameeh, V. , Fanaee, H. R. , & Hezarjeribi, E. (2016). Univariate stability analysis of genotype× environment interaction of oilseed rape seed yield. Acta Universitatis Agriculturae et Silviculturae Mendelianae Brunensis, 64, 1625–1634.
-
- Dehghani, H. , Omidi, H. , & Sabaghnia, N. J. A. J. (2008). Graphic analysis of trait relations of rapeseed using the biplot method. Agronomy Journal, 100, 1443–1449.
-
- Escobar, M. , Berti, M. , Matus, I. , Tapia, M. , & Johnson, B. (2011). Genotype× environment interaction in canola (Brassica napus L.) seed yield in Chile. Chilean Journal of Agricultural Research, 71, 175–186.
-
- FAO . (2018). Food and Agriculture Organization of the United Nations, Food and Agricultural Commodities Production . http://www.fao.org/statistics/en
LinkOut - more resources
Full Text Sources
Miscellaneous

