Assembly and disassembly of branched ubiquitin chains
- PMID: 37325469
- PMCID: PMC10267395
- DOI: 10.3389/fmolb.2023.1197272
Assembly and disassembly of branched ubiquitin chains
Abstract
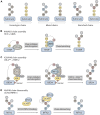
Protein ubiquitylation is an essential post-translational modification that regulates nearly all aspects of eukaryotic cell biology. A diverse collection of ubiquitylation signals, including an extensive repertoire of polymeric ubiquitin chains, leads to a range of different functional outcomes for the target protein. Recent studies have shown that ubiquitin chains can be branched and that branched chains have a direct impact on the stability or the activity of the target proteins they are attached to. In this mini review, we discuss the mechanisms that control the assembly and disassembly of branched chains by the enzymes of the ubiquitylation and deubiquitylation machinery. Existing knowledge regarding the activities of chain branching ubiquitin ligases and the deubiquitylases responsible for cleaving branched chains is summarized. We also highlight new findings concerning the formation of branched chains in response to small molecules that induce the degradation of otherwise stable proteins and examine the selective debranching of heterotypic chains by the proteasome-bound deubiquitylase UCH37.
Keywords: branched ubiquitin chain; proteasome; protein degradation; ubiquitin; ubiquitylation (ubiquitination).
Copyright © 2023 Gregor, Xu and French.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

