Salmonella Enteritidis activates inflammatory storm via SPI-1 and SPI-2 to promote intracellular proliferation and bacterial virulence
- PMID: 37325511
- PMCID: PMC10266283
- DOI: 10.3389/fcimb.2023.1158888
Salmonella Enteritidis activates inflammatory storm via SPI-1 and SPI-2 to promote intracellular proliferation and bacterial virulence
Abstract
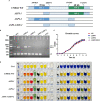
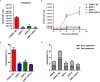
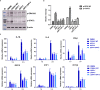
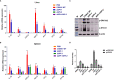
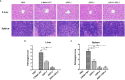
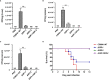
Salmonella Enteritidis is an important intracellular pathogen, which can cause gastroenteritis in humans and animals and threaten life and health. S. Enteritidis proliferates in host macrophages to establish systemic infection. In this study, we evaluated the effects of Salmonella pathogenicity island-1 (SPI-1) and SPI-2 to S. Enteritidis virulence in vitro and in vivo, as well as the host inflammatory pathways affected by SPI-1 and SPI-2. Our results show that S. Enteritidis SPI-1 and SPI-2 contributed to bacterial invasion and proliferation in RAW264.7 macrophages, and induced cytotoxicity and cellular apoptosis of these cells. S. Enteritidis infection induced multiple inflammatory responses, including mitogen-activated protein kinase (ERK-mediated) and Janus kinase-signal transducer and activator of transcript (STAT) (STAT2-mediated) pathways. Both SPI-1 and SPI-2 were necessary to induce robust inflammatory responses and ERK/STAT2 phosphorylation in macrophages. In a mouse infection model, both SPIs, especially SPI-2, resulted in significant production of inflammatory cytokines and various interferon-stimulated genes in the liver and spleen. Activation of the ERK- and STAT2-mediated cytokine storm was largely affected by SPI-2. S. Enteritidis ΔSPI-1-infected mice displayed moderate histopathological damage and drastically reduced bacterial loads in tissues, whereas only slight damage and no bacteria were observed in ΔSPI-2- and ΔSPI-1/SPI-2-infected mice. A survival assay showed that ΔSPI-1 mutant mice maintained a medium level of virulence, while SPI-2 plays a decisive role in bacterial virulence. Collectively, our findings indicate that both SPIs, especially SPI-2, profoundly contributed to S. Enteritidis intracellular localization and virulence by activating multiple inflammatory pathways.
Keywords: Inflammatory pathway; Salmonella Pathogenicity Island; bacterial colonization; macrophage; mouse infection model.
Copyright © 2023 Xiong, Song, Chen, Jiao and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Identification of a novel gene in ROD9 island of Salmonella Enteritidis involved in the alteration of virulence-associated genes expression.Virulence. 2018 Jan 1;9(1):348-362. doi: 10.1080/21505594.2017.1392428. Virulence. 2018. PMID: 29130383 Free PMC article.
-
Modulation of Salmonella virulence by a novel SPI-2 injectisome effector that interacts with the dystrophin-associated protein complex.mBio. 2024 Jul 17;15(7):e0112824. doi: 10.1128/mbio.01128-24. Epub 2024 Jun 21. mBio. 2024. PMID: 38904384 Free PMC article.
-
Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens.BMC Microbiol. 2009 Dec 19;9:268. doi: 10.1186/1471-2180-9-268. BMC Microbiol. 2009. PMID: 20021686 Free PMC article.
-
The role of host cell death in Salmonella infections.Curr Top Microbiol Immunol. 2005;289:131-50. doi: 10.1007/3-540-27320-4_6. Curr Top Microbiol Immunol. 2005. PMID: 15791954 Review.
-
[Molecular mechanism for pathogenicity of Salmonella sp].Rev Latinoam Microbiol. 2005 Jan-Jun;47(1-2):25-42. Rev Latinoam Microbiol. 2005. PMID: 17061544 Review. Spanish.
Cited by
-
Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes.Antibiotics (Basel). 2023 Dec 28;13(1):30. doi: 10.3390/antibiotics13010030. Antibiotics (Basel). 2023. PMID: 38247590 Free PMC article.
-
PipC affects the virulence of Salmonella enterica serovar Enteritidis and its deletion strain provides effective immune protection in mice.Front Microbiol. 2025 Jun 24;16:1631008. doi: 10.3389/fmicb.2025.1631008. eCollection 2025. Front Microbiol. 2025. PMID: 40630190 Free PMC article.
-
Whole-genome sequencing-based characterization of Salmonella enterica Serovar Enteritidis and Kentucky isolated from laying hens in northwest of Iran, 2022-2023.Gut Pathog. 2025 Jan 16;17(1):2. doi: 10.1186/s13099-025-00679-3. Gut Pathog. 2025. PMID: 39819347 Free PMC article.
-
Living hybrid material based on probiotic with photothermal properties inhibits PD-L1 expression after tumouricidal photothermal therapy.Biomater Transl. 2025 Mar 25;6(1):73-84. doi: 10.12336/biomatertransl.2025.01.006. eCollection 2025. Biomater Transl. 2025. PMID: 40313568 Free PMC article.
-
Regulatory role of the Cpx ESR in bacterial behaviours.Virulence. 2024 Dec;15(1):2404951. doi: 10.1080/21505594.2024.2404951. Epub 2024 Sep 18. Virulence. 2024. PMID: 39292643 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention (2023) Salmonella. Available at: https://www.cdc.gov/salmonella/index.html (Accessed 2, 2023).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

