Antagonistic transcriptome profile reveals potential mechanisms of action on Xanthomonas oryzae pv. oryzicola by the cell-free supernatants of Bacillus velezensis 504, a versatile plant probiotic bacterium
- PMID: 37325518
- PMCID: PMC10265122
- DOI: 10.3389/fcimb.2023.1175446
Antagonistic transcriptome profile reveals potential mechanisms of action on Xanthomonas oryzae pv. oryzicola by the cell-free supernatants of Bacillus velezensis 504, a versatile plant probiotic bacterium
Abstract
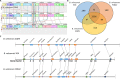
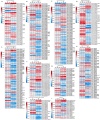
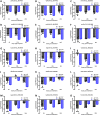
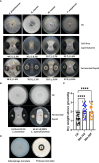
Bacterial leaf streak (BLS) of rice is a severe disease caused by the bacterial pathogen Xanthomonas oryzae pv. oryzicola (Xoc) that has gradually become the fourth major disease on rice in some rice-growing regions in southern China. Previously, we isolated a Bacillus velezensis strain 504 that exhibited apparent antagonistic activity against the Xoc wild-type strain RS105, and found that B. velezensis 504 was a potential biocontrol agent for BLS. However, the underlying mechanisms of antagonism and biocontrol are not completely understood. Here we mine the genomic data of B. velezensis 504, and the comparative transcriptomic data of Xoc RS105 treated by the cell-free supernatants (CFSs) of B. velezensis 504 to define differentially expressed genes (DEGs). We show that B. velezensis 504 shares over 89% conserved genes with FZB42 and SQR9, two representative model strains of B. velezensis, but 504 is more closely related to FZB42 than SQR9, as well as B. velezensis 504 possesses the secondary metabolite gene clusters encoding the essential anti-Xoc agents difficidin and bacilysin. We conclude that approximately 77% of Xoc RS105 coding sequences are differentially expressed by the CFSs of B. velezensis 504, which significantly downregulates genes involved in signal transduction, oxidative phosphorylation, transmembrane transport, cell motility, cell division, DNA translation, and five physiological metabolisms, as well as depresses an additional set of virulence-associated genes encoding the type III secretion, type II secretion system, type VI secretion system, type IV pilus, lipopolysaccharides and exopolysaccharides. We also show that B. velezensis 504 is a potential biocontrol agent for bacterial blight of rice exhibiting relative control efficiencies over 70% on two susceptible cultivars, and can efficiently antagonize against some important plant pathogenic fungi including Colletotrichum siamense and C. australisinense that are thought to be the two dominant pathogenic species causing leaf anthracnose of rubber tree in Hainan province of China. B. velezensis 504 also harbors some characteristics of plant growth-promoting rhizobacterium such as secreting protease and siderophore, and stimulating plant growth. This study reveals the potential biocontrol mechanisms of B. velezensis against BLS, and also suggests that B. velezensis 504 is a versatile plant probiotic bacterium.
Keywords: Bacillus velezensis; Xanthomonas oryzae pv. oryzicola; antagonism mechanism; biocontrol agent; transcriptome profiling.
Copyright © 2023 Zhou, Tu, Fu, Chen, Wang, Fang, Yan, Cheng, Zhang, Zhu, Yin, Xiao, Zou and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
A New Biocontrol Agent Bacillus velezensis SF334 against Rubber Tree Fungal Leaf Anthracnose and Its Genome Analysis of Versatile Plant Probiotic Traits.J Fungi (Basel). 2024 Feb 17;10(2):158. doi: 10.3390/jof10020158. J Fungi (Basel). 2024. PMID: 38392830 Free PMC article.
-
Comparative transcriptomic profiling of the two-stage response of rice to Xanthomonas oryzae pv. oryzicola interaction with two different pathogenic strains.BMC Plant Biol. 2024 Apr 29;24(1):347. doi: 10.1186/s12870-024-05060-1. BMC Plant Biol. 2024. PMID: 38684939 Free PMC article.
-
AvrXa7-Xa7 mediated defense in rice can be suppressed by transcriptional activator-like effectors TAL6 and TAL11a from Xanthomonas oryzae pv. oryzicola.Mol Plant Microbe Interact. 2014 Sep;27(9):983-95. doi: 10.1094/MPMI-09-13-0279-R. Mol Plant Microbe Interact. 2014. Retraction in: Mol Plant Microbe Interact. 2014 Dec;27(12):1413. PMID: 25105804 Retracted.
-
Bacillus velezensis: a versatile ally in the battle against phytopathogens-insights and prospects.Appl Microbiol Biotechnol. 2024 Aug 15;108(1):439. doi: 10.1007/s00253-024-13255-7. Appl Microbiol Biotechnol. 2024. PMID: 39145847 Free PMC article. Review.
-
Biological control of bacterial leaf blight (BLB) in rice-A sustainable approach.Heliyon. 2025 Jan 8;11(2):e41769. doi: 10.1016/j.heliyon.2025.e41769. eCollection 2025 Jan 30. Heliyon. 2025. PMID: 39872461 Free PMC article. Review.
Cited by
-
A New Biocontrol Agent Bacillus velezensis SF334 against Rubber Tree Fungal Leaf Anthracnose and Its Genome Analysis of Versatile Plant Probiotic Traits.J Fungi (Basel). 2024 Feb 17;10(2):158. doi: 10.3390/jof10020158. J Fungi (Basel). 2024. PMID: 38392830 Free PMC article.
References
-
- Borriss R., Wu H., Gao X. (2019). Secondary metabolites of the plant growth promoting model rhizobacterium Bacillus velezensis FZB42 are involved in direct suppression of plant pathogens and in stimulation of plant-induced systemic resistance. Secondary. Metabol. Plant Growth Promoting. Rhizomicroorg.: Discovery Appl., 147–168. doi: 10.1007/978-981-13-5862-3_8 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

