Single-cell and spatial transcriptome analysis reveals the cellular heterogeneity of liver metastatic colorectal cancer
- PMID: 37327339
- PMCID: PMC10275599
- DOI: 10.1126/sciadv.adf5464
Single-cell and spatial transcriptome analysis reveals the cellular heterogeneity of liver metastatic colorectal cancer
Abstract
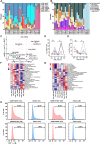
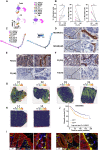
In this study, we comprehensively charted the cellular landscape of colorectal cancer (CRC) and well-matched liver metastatic CRC using single-cell and spatial transcriptome RNA sequencing. We generated 41,892 CD45- nonimmune cells and 196,473 CD45+ immune cells from 27 samples of six CRC patients, and found that CD8_CXCL13 and CD4_CXCL13 subsets increased significantly in liver metastatic samples that exhibited high proliferation ability and tumor-activating characterization, contributing to better prognosis of patients. Distinct fibroblast profiles were observed in primary and liver metastatic tumors. F3+ fibroblasts enriched in primary tumors contributed to worse overall survival by expressing protumor factors. However, MCAM+ fibroblasts enriched in liver metastatic tumors might promote generation of CD8_CXCL13 cells through Notch signaling. In summary, we extensively analyzed the transcriptional differences of cell atlas between primary and liver metastatic tumors of CRC by single-cell and spatial transcriptome RNA sequencing, providing different dimensions of the development of liver metastasis in CRC.
Figures







References
-
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). - PubMed
-
- R. L. Siegel, K. D. Miller, A. Jemal, Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). - PubMed
-
- D. G. DeNardo, M. Johansson, L. M. Coussens, Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev. 27, 11–18 (2008). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

