Analysis of gut microbiota in patients with Williams-Beuren Syndrome reveals dysbiosis linked to clinical manifestations
- PMID: 37328513
- PMCID: PMC10275996
- DOI: 10.1038/s41598-023-36704-1
Analysis of gut microbiota in patients with Williams-Beuren Syndrome reveals dysbiosis linked to clinical manifestations
Abstract
Williams-Beuren syndrome (WBS) is a multisystem genetic disease caused by the deletion of a region of 1.5-1.8 Mb on chromosome 7q11.23. The elastin gene seems to account for several comorbidities and distinct clinical features such including cardiovascular disease, connective tissue abnormalities, growth retardation, and gastrointestinal (GI) symptoms. Increasing evidence points to alterations in gut microbiota composition as a primary or secondary cause of some GI or extra-intestinal characteristics. In this study, we performed the first exploratory analysis of gut microbiota in WBS patients compared to healthy subjects (CTRLs) using 16S rRNA amplicon sequencing, by investigating the gut dysbiosis in relation to diseases and comorbidities. We found that patients with WBS have significant dysbiosis compared to age-matched CTRLs, characterized by an increase in proinflammatory bacteria such as Pseudomonas, Gluconacetobacter and Eggerthella, and a reduction of anti-inflammatory bacteria including Akkermansia and Bifidobacterium. Microbial biomarkers associated with weight gain, GI symptoms and hypertension were identified. Gut microbiota profiling could represent a new tool that characterise intestinal dysbiosis to complement the clinical management of these patients. In particular, the administration of microbial-based treatments, alongside traditional therapies, could help in reducing or preventing the burden of these symptoms and improve the quality of life of these patients.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

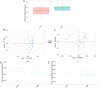
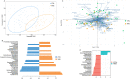


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

