A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity
- PMID: 37328517
- PMCID: PMC10275998
- DOI: 10.1038/s41467-023-39254-2
A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity
Abstract
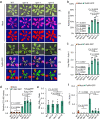
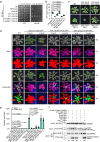
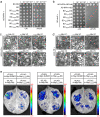
NONEXPRESSER OF PATHOGENESIS-RELATED GENES 1 (NPR1) is the master regulator of salicylic acid-mediated basal and systemic acquired resistance in plants. Here, we report that NPR1 plays a pivotal role in restricting compatible infection by turnip mosaic virus, a member of the largest plant RNA virus genus Potyvirus, and that such resistance is counteracted by NUCLEAR INCLUSION B (NIb), the viral RNA-dependent RNA polymerase. We demonstrate that NIb binds to the SUMO-interacting motif 3 (SIM3) of NPR1 to prevent SUMO3 interaction and sumoylation, while sumoylation of NIb by SUMO3 is not essential but can intensify the NIb-NPR1 interaction. We discover that the interaction also impedes the phosphorylation of NPR1 at Ser11/Ser15. Moreover, we show that targeting NPR1 SIM3 is a conserved ability of NIb from diverse potyviruses. These data reveal a molecular "arms race" by which potyviruses deploy NIb to suppress NPR1-mediated resistance through disrupting NPR1 sumoylation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

