Identification of molecular mechanisms causing skin lesions of cutaneous leishmaniasis using weighted gene coexpression network analysis (WGCNA)
- PMID: 37330553
- PMCID: PMC10276835
- DOI: 10.1038/s41598-023-35868-0
Identification of molecular mechanisms causing skin lesions of cutaneous leishmaniasis using weighted gene coexpression network analysis (WGCNA)
Abstract
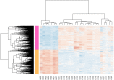
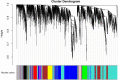
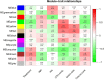
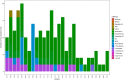
Leishmaniasis is an infectious disease, caused by a protozoan parasite. Its most common form is cutaneous leishmaniasis, which leaves scars on exposed body parts from bites by infected female phlebotomine sandflies. Approximately 50% of cases of cutaneous leishmaniasis fail to respond to standard treatments, creating slow-healing wounds which cause permanent scars on the skin. We performed a joint bioinformatics analysis to identify differentially expressed genes (DEGs) in healthy skin biopsies and Leishmania cutaneous wounds. DEGs and WGCNA modules were analyzed based on the Gene Ontology function, and the Cytoscape software. Among almost 16,600 genes that had significant expression changes on the skin surrounding Leishmania wounds, WGCNA determined that one of the modules, with 456 genes, has the strongest correlation with the size of the wounds. Functional enrichment analysis indicated that this module includes three gene groups with significant expression changes. These produce tissue-damaging cytokines or disrupt the production and activation of collagen, fibrin proteins, and the extracellular matrix, causing skin wounds or preventing them from healing. The hub genes of these groups are OAS1, SERPINH1, and FBLN1 respectively. This information can provide new ways to deal with unwanted and harmful effects of cutaneous leishmaniasis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Integrated analysis of lncRNA and mRNA expression profiles in cutaneous leishmaniasis lesions caused by Leishmania tropica.Front Cell Infect Microbiol. 2024 Nov 21;14:1416925. doi: 10.3389/fcimb.2024.1416925. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39639867 Free PMC article.
-
[Defining the Molecular Signal Pathways and Upstream Regulators in Cutaneous Leishmaniasis with Transcriptomic Data Approach].Mikrobiyol Bul. 2021 Jan;55(1):67-80. doi: 10.5578/mb.20092. Mikrobiyol Bul. 2021. PMID: 33590982 Turkish.
-
Transcriptional Analysis of Human Skin Lesions Identifies Tryptophan-2,3-Deoxygenase as a Restriction Factor for Cutaneous Leishmania.Front Cell Infect Microbiol. 2019 Oct 4;9:338. doi: 10.3389/fcimb.2019.00338. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31637219 Free PMC article.
-
The Impact of Neutrophil Recruitment to the Skin on the Pathology Induced by Leishmania Infection.Front Immunol. 2021 Mar 1;12:649348. doi: 10.3389/fimmu.2021.649348. eCollection 2021. Front Immunol. 2021. PMID: 33732265 Free PMC article. Review.
-
Cutaneous Manifestations of Human and Murine Leishmaniasis.Int J Mol Sci. 2017 Jun 18;18(6):1296. doi: 10.3390/ijms18061296. Int J Mol Sci. 2017. PMID: 28629171 Free PMC article. Review.
Cited by
-
Overview of Research on Leishmaniasis in Africa: Current Status, Diagnosis, Therapeutics, and Recent Advances Using By-Products of the Sargassaceae Family.Pharmaceuticals (Basel). 2024 Apr 18;17(4):523. doi: 10.3390/ph17040523. Pharmaceuticals (Basel). 2024. PMID: 38675483 Free PMC article.
References
-
- Bhar A. The application of next generation sequencing technology in medical diagnostics: A perspective. ProcIndian Natl. Sci. Acad. 2022;88(4):592–600. doi: 10.1007/s43538-022-00098-x. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

