ZEB1 Transcriptionally Activates PHGDH to Facilitate Carcinogenesis and Progression of HCC
- PMID: 37331567
- PMCID: PMC10469392
- DOI: 10.1016/j.jcmgh.2023.06.006
ZEB1 Transcriptionally Activates PHGDH to Facilitate Carcinogenesis and Progression of HCC
Abstract
Background & aims: Phosphoglycerate dehydrogenase (PHGDH), the rate-limiting enzyme of the de novo serine synthesis pathway (SSP), has been implicated in the carcinogenesis and metastasis of hepatocellular carcinoma (HCC) because of its excessive expression and promotion of SSP. In previous experiments we found that SSP flux was diminished by knockdown of zinc finger E-box binding homeobox 1 (ZEB1), a stimulator of HCC metastasis, but the underlying mechanism remains largely unknown. Here, we aimed to determine how SSP flux is regulated by ZEB1 and the contribution of such regulation to carcinogenesis and progression of HCC.
Methods: We used genetic mice with Zeb1 knockout in liver specifically to determine whether Zeb1 deficiency impacts HCC induced by the carcinogen diethylnitrosamine plus CCl4. We explored the regulatory mechanism of ZEB1 in SSP flux using uniformly-labeled [13C]-glucose tracing analyses, liquid chromatography-mass spectrometry, real-time quantitative polymerase chain reaction, luciferase report assay, and chromatin immunoprecipitation assay. We determined the contribution of the ZEB1-PHGDH regulatory axis to carcinogenesis and metastasis of HCC by cell counting assay, methyl thiazolyl tetrazolium (MTT) assay, scratch wound assay, Transwell assay, and soft agar assay in vitro, orthotopic xenograft, bioluminescence, and H&E assays in vivo. We investigated the clinical relevance of ZEB1 and PHGDH by analyzing publicly available data sets and 48 pairs of HCC clinical specimens.
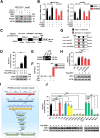
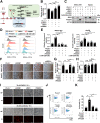
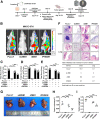
Results: We identified that ZEB1 activates PHGDH transcription by binding to a nonclassic binding site within its promoter region. Up-regulated PHGDH augments SSP flux to enable HCC cells to be more invasive, proliferative, and resistant to reactive oxygen species and sorafenib. Orthotopic xenograft and bioluminescence assays have shown that ZEB1 deficiency significantly impairs the tumorigenesis and metastasis of HCC, and such impairments can be rescued to a large extent by exogenous expression of PHGDH. These results were confirmed by the observation that conditional knockout of ZEB1 in mouse liver dramatically impedes carcinogenesis and progression of HCC induced by diethylnitrosamine/CCl4, as well as PHGDH expression. In addition, analysis of The Cancer Genome Atlas database and clinical HCC samples showed that the ZEB1-PHGDH regulatory axis predicts poor prognosis of HCC.
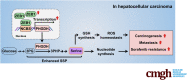
Conclusions: ZEB1 plays a crucial role in stimulating carcinogenesis and progression of HCC by activating PHGDH transcription and subsequent SSP flux, deepening our knowledge of ZEB1 as a transcriptional factor in fostering the development of HCC via reprogramming the metabolic pathway.
Keywords: De Novo Serine Synthesis Pathway; Hepatocellular Carcinoma; Metabolic Reprogramming; Tumor Metastasis.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
ZEB1 Fuels Serine Metabolism to Promote HCC Metastasis.Cell Mol Gastroenterol Hepatol. 2023;16(4):646-647. doi: 10.1016/j.jcmgh.2023.07.002. Epub 2023 Jul 28. Cell Mol Gastroenterol Hepatol. 2023. PMID: 37517803 Free PMC article. No abstract available.
References
-
- Craig A.J., von Felden J., Garcia-Lezana T., et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139–152. - PubMed
-
- Vogel A., Meyer T., Sapisochin G., et al. Hepatocellular carcinoma. Lancet. 2022;400:1345–1362. - PubMed
-
- Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

