Lipid biosynthesis perturbation impairs endoplasmic reticulum-associated degradation
- PMID: 37331602
- PMCID: PMC10372827
- DOI: 10.1016/j.jbc.2023.104939
Lipid biosynthesis perturbation impairs endoplasmic reticulum-associated degradation
Abstract
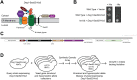
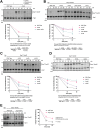
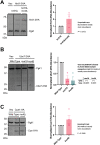
The relationship between lipid homeostasis and protein homeostasis (proteostasis) is complex and remains incompletely understood. We conducted a screen for genes required for efficient degradation of Deg1-Sec62, a model aberrant translocon-associated substrate of the endoplasmic reticulum (ER) ubiquitin ligase Hrd1, in Saccharomyces cerevisiae. This screen revealed that INO4 is required for efficient Deg1-Sec62 degradation. INO4 encodes one subunit of the Ino2/Ino4 heterodimeric transcription factor, which regulates expression of genes required for lipid biosynthesis. Deg1-Sec62 degradation was also impaired by mutation of genes encoding several enzymes mediating phospholipid and sterol biosynthesis. The degradation defect in ino4Δ yeast was rescued by supplementation with metabolites whose synthesis and uptake are mediated by Ino2/Ino4 targets. Stabilization of a panel of substrates of the Hrd1 and Doa10 ER ubiquitin ligases by INO4 deletion indicates ER protein quality control is generally sensitive to perturbed lipid homeostasis. Loss of INO4 sensitized yeast to proteotoxic stress, suggesting a broad requirement for lipid homeostasis in maintaining proteostasis. A better understanding of the dynamic relationship between lipid homeostasis and proteostasis may lead to improved understanding and treatment of several human diseases associated with altered lipid biosynthesis.
Keywords: Doa10; ER quality control; Hrd1; Saccharomyces cerevisiae; endoplasmic reticulum-associated degradation (ERAD); phospholipid metabolism; protein degradation; sterol; translocon quality control; yeast genetics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Berner N., Reutter K.R., Wolf D.H. Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path. Annu. Rev. Biochem. 2018;87:751–782. - PubMed
-
- Plemper R.K., Bordallo J., Deak P.M., Taxis C., Hitt R., Wolf D.H. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci. 1999;112:4123–4134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

