T cells in health and disease
- PMID: 37332039
- PMCID: PMC10277291
- DOI: 10.1038/s41392-023-01471-y
T cells in health and disease
Abstract
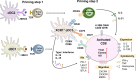
T cells are crucial for immune functions to maintain health and prevent disease. T cell development occurs in a stepwise process in the thymus and mainly generates CD4+ and CD8+ T cell subsets. Upon antigen stimulation, naïve T cells differentiate into CD4+ helper and CD8+ cytotoxic effector and memory cells, mediating direct killing, diverse immune regulatory function, and long-term protection. In response to acute and chronic infections and tumors, T cells adopt distinct differentiation trajectories and develop into a range of heterogeneous populations with various phenotype, differentiation potential, and functionality under precise and elaborate regulations of transcriptional and epigenetic programs. Abnormal T-cell immunity can initiate and promote the pathogenesis of autoimmune diseases. In this review, we summarize the current understanding of T cell development, CD4+ and CD8+ T cell classification, and differentiation in physiological settings. We further elaborate the heterogeneity, differentiation, functionality, and regulation network of CD4+ and CD8+ T cells in infectious disease, chronic infection and tumor, and autoimmune disease, highlighting the exhausted CD8+ T cell differentiation trajectory, CD4+ T cell helper function, T cell contributions to immunotherapy and autoimmune pathogenesis. We also discuss the development and function of γδ T cells in tissue surveillance, infection, and tumor immunity. Finally, we summarized current T-cell-based immunotherapies in both cancer and autoimmune diseases, with an emphasis on their clinical applications. A better understanding of T cell immunity provides insight into developing novel prophylactic and therapeutic strategies in human diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

