Selective polypeptide ligand binding to the extracellular surface of the transmembrane domains of the class B GPCRs GLP-1R and GCGR
- PMID: 37332600
- PMCID: PMC10276138
- DOI: 10.1016/j.isci.2023.106918
Selective polypeptide ligand binding to the extracellular surface of the transmembrane domains of the class B GPCRs GLP-1R and GCGR
Abstract
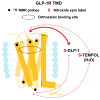
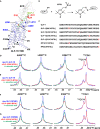
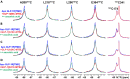
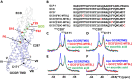
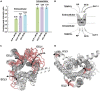
Crystal and cryo-EM structures of the glucagon-like peptide-1 receptor (GLP-1R) and glucagon receptor (GCGR) bound with their peptide ligands have been obtained with full-length constructs, indicating that the extracellular domain (ECD) is indispensable for specific ligand binding. This article complements these data with studies of ligand recognition of the two receptors in solution. Paramagnetic NMR relaxation enhancement measurements using dual labeling with fluorine-19 probes on the receptor and nitroxide spin labels on the peptide ligands provided new insights. The glucagon-like peptide-1 (GLP-1) was found to interact with GLP-1R by selective binding to the extracellular surface. The ligand selectivity toward the extracellular surface of the receptor was preserved in the transmembrane domain (TMD) devoid of the ECD. The dual labeling approach further provided evidence of cross-reactivity of GLP-1R and GCGR with glucagon and GLP-1, respectively, which is of interest in the context of medical treatments using combinations of the two polypeptides.
Keywords: Biological sciences; molecular biology; molecular interaction; molecular structure.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Graaf C.d., Donnelly D., Wootten D., Lau J., Sexton P.M., Miller L.J., Ahn J.M., Liao J., Fletcher M.M., Yang D., et al. Glucagon-like peptide-1 and its class B G protein-coupled receptors: a long march to therapeutic successes. Pharmacol. Rev. 2016;68:954–1013. doi: 10.1124/pr.115.011395. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

