Differentially expressed miRNA profiles of serum-derived exosomes in patients with sudden sensorineural hearing loss
- PMID: 37332997
- PMCID: PMC10273844
- DOI: 10.3389/fneur.2023.1177988
Differentially expressed miRNA profiles of serum-derived exosomes in patients with sudden sensorineural hearing loss
Abstract
Objectives: This study aimed to compare the expressed microRNA (miRNA) profiles of serum-derived exosomes of patients with sudden sensorineural hearing loss (SSNHL) and normal hearing controls to identify exosomal miRNAs that may be associated with SSNHL or serve as biomarkers for SSNHL.
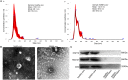
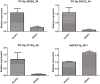
Methods: Peripheral venous blood of patients with SSNHL and healthy controls was collected to isolate exosomes. Nanoparticle tracking analysis, transmission electron microscopy, and Western blotting were used to identify the isolated exosomes, after which total RNA was extracted and used for miRNA transcriptome sequencing. Differentially expressed miRNAs (DE-miRNAs) were identified based on the thresholds of P < 0.05 and |log2fold change| > 1 and subjected to functional analyses. Finally, four exosomal DE-miRNAs, including PC-5p-38556_39, PC-5p-29163_54, PC-5p-31742_49, and hsa-miR-93-3p_R+1, were chosen for validation using quantitative real-time polymerase chain reaction (RT-qPCR).
Results: Exosomes were isolated from serum and identified based on particle size, morphological examination, and expression of exosome-marker proteins. A total of 18 exosomal DE-miRNAs, including three upregulated and 15 downregulated miRNAs, were found in SSNHL cases. Gene ontology (GO) functional annotation analysis revealed that target genes in the top 20 terms were mainly related to "protein binding," "metal ion binding," "ATP binding," and "intracellular signal transduction." Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that these target genes were functionally enriched in the "Ras," "Hippo," "cGMP-PKG," and "AMPK signaling pathways." The expression levels of PC-5p-38556_39 and PC-5p-29163_54 were significantly downregulated and that of miR-93-3p_R+1 was highly upregulated in SSNHL. Consequently, the consistency rate between sequencing and RT-qPCR was 75% and sequencing results were highly reliable.
Conclusion: This study identified 18 exosomal DE-miRNAs, including PC-5p-38556_39, PC-5p-29163_54, and miR-93-3p, which may be closely related to SSNHL pathogenesis or serve as biomarkers for SSNHL.
Keywords: biomarkers; exosomes; miRNA; miRNA transcriptome sequencing; sudden sensorineural hearing loss.
Copyright © 2023 Zhang, Ma, Yang, Ke, Sun, Yang, Kuang, Li and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Expression profile of serum-related exosomal miRNAs from parathyroid tumor.Endocrine. 2021 Apr;72(1):239-248. doi: 10.1007/s12020-020-02535-7. Epub 2020 Nov 8. Endocrine. 2021. PMID: 33161496
-
Prognostic implication of downregulated exosomal miRNAs in patients with sepsis: a cross-sectional study with bioinformatics analysis.J Intensive Care. 2023 Aug 3;11(1):35. doi: 10.1186/s40560-023-00683-2. J Intensive Care. 2023. PMID: 37537685 Free PMC article.
-
Small RNA sequencing and profiling of serum-derived exosomes from African swine fever virus-infected pigs.J Anim Sci. 2023 Jan 3;101:skac400. doi: 10.1093/jas/skac400. J Anim Sci. 2023. PMID: 36478238 Free PMC article.
-
Differential expression of exosomal microRNAs in fresh and senescent apheresis platelet concentrates.Platelets. 2022 Nov 17;33(8):1260-1269. doi: 10.1080/09537104.2022.2108541. Epub 2022 Aug 14. Platelets. 2022. PMID: 35968647
-
Analysis of MicroRNA Signature Differentially Expressed in Pancreatic Islet Cells Treated with Pancreatic Cancer-Derived Exosomes.Int J Mol Sci. 2023 Sep 19;24(18):14301. doi: 10.3390/ijms241814301. Int J Mol Sci. 2023. PMID: 37762604 Free PMC article. Review.
Cited by
-
NcRNA: key and potential in hearing loss.Front Neurosci. 2024 Jan 17;17:1333131. doi: 10.3389/fnins.2023.1333131. eCollection 2023. Front Neurosci. 2024. PMID: 38298898 Free PMC article. Review.
-
Pathological noise exposure results in elevated levels of extracellular vesicles enriched with Hsp70 in the plasma.J Mol Med (Berl). 2025 Aug 16. doi: 10.1007/s00109-025-02584-3. Online ahead of print. J Mol Med (Berl). 2025. PMID: 40817924
-
Serum and Plasma miRNA Expression Levels in Sudden Sensorineural Hearing Loss.Int J Mol Sci. 2025 Jan 31;26(3):1245. doi: 10.3390/ijms26031245. Int J Mol Sci. 2025. PMID: 39941013 Free PMC article.
References
LinkOut - more resources
Full Text Sources

