This is a preprint.
Myeloid cell influx into the colonic epithelium is associated with disease severity and non-response to anti-Tumor Necrosis Factor Therapy in patients with Ulcerative Colitis
- PMID: 37333091
- PMCID: PMC10274630
- DOI: 10.1101/2023.06.02.542863
Myeloid cell influx into the colonic epithelium is associated with disease severity and non-response to anti-Tumor Necrosis Factor Therapy in patients with Ulcerative Colitis
Abstract
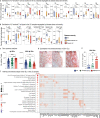
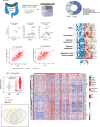
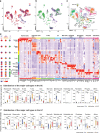
Ulcerative colitis (UC) is an idiopathic chronic inflammatory disease of the colon with sharply rising global prevalence. Dysfunctional epithelial compartment (EC) dynamics are implicated in UC pathogenesis although EC-specific studies are sparse. Applying orthogonal high-dimensional EC profiling to a Primary Cohort (PC; n=222), we detail major epithelial and immune cell perturbations in active UC. Prominently, reduced frequencies of mature BEST4+OTOP2+ absorptive and BEST2+WFDC2+ secretory epithelial enterocytes were associated with the replacement of homeostatic, resident TRDC+KLRD1+HOPX+ γδ+ T cells with RORA+CCL20+S100A4+ TH17 cells and the influx of inflammatory myeloid cells. The EC transcriptome (exemplified by S100A8, HIF1A, TREM1, CXCR1) correlated with clinical, endoscopic, and histological severity of UC in an independent validation cohort (n=649). Furthermore, therapeutic relevance of the observed cellular and transcriptomic changes was investigated in 3 additional published UC cohorts (n=23, 48 and 204 respectively) to reveal that non-response to anti-Tumor Necrosis Factor (anti-TNF) therapy was associated with EC related myeloid cell perturbations. Altogether, these data provide high resolution mapping of the EC to facilitate therapeutic decision-making and personalization of therapy in patients with UC.
Keywords: RNA sequencing; Ulcerative colitis; anti-TNF treatment response; intraepithelial compartment; spatial transcriptomics.
Conflict of interest statement
Competing interests: SM reports receiving research grants from Genentech and Takeda; receiving payment for lectures from Takeda, Genentech, Morphic; and receiving consulting fees from Takeda, Morphic, Ferring and Arena Pharmaceuticals. JM reports receiving consulting fees from Janssen Pharmaceuticals. JFC reports receiving research grants from AbbVie, Janssen Pharmaceuticals, Takeda and Bristol Myers Squibb; receiving payment for lectures from AbbVie, and Takeda; receiving consulting fees from AbbVie, Amgen, AnaptysBio, Allergan, Arena Pharmaceuticals, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Celltrion, Eli Lilly, Ferring Pharmaceuticals, Galmed Research, Glaxo Smith Kline, Genentech (Roche), Janssen Pharmaceuticals, Kaleido Biosciences, Immunic, Invea, Iterative Scopes, Merck, Landos, Microba Life Science, Novartis, Otsuka Pharmaceutical, Pfizer, Protagonist Therapeutics, Prometheus, Sanofi, Seres, Takeda, Teva, TiGenix, Vifor; and hold stock options in Intestinal Biotech Development. JM reports receiving consulting fees from Janssen Pharmaceuticals. RCU has served as an advisory board member or consultant for AbbVie, Bristol Myers Squibb, Celltrion, Janssen, Pfizer, Roivant, and Takeda; research support from AbbVie, Boehringer Ingelheim, Eli Lily, and Pfizer. MK has served as a consultant for AbbVie, Pfizer, Bristol Myers Squibb, Fresenius, GoodRx.
Figures












References
-
- Liang S., et al. Nutrient Sensing by the Intestinal Epithelium Orchestrates Mucosal Antimicrobial Defense via Translational Control of Hes1. Cell Host Microbe 25, 706–718 e707 (2019). - PubMed
-
- Kopf A. & Sixt M. Gut Homeostasis: Active Migration of Intestinal Epithelial Cells in Tissue Renewal. Curr Biol 29, R1091–R1093 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
