This is a preprint.
Beyond antibiotic resistance: the whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria
- PMID: 37333137
- PMCID: PMC10274678
- DOI: 10.1101/2023.06.02.543512
Beyond antibiotic resistance: the whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria
Update in
-
Beyond antibiotic resistance: The whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria.Cell Chem Biol. 2024 Apr 18;31(4):669-682.e7. doi: 10.1016/j.chembiol.2023.12.020. Epub 2024 Jan 23. Cell Chem Biol. 2024. PMID: 38266648 Free PMC article.
Abstract
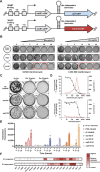
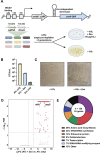
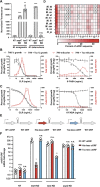
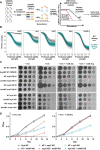
Pathogenic mycobacteria are a significant cause of morbidity and mortality worldwide. These bacteria are highly intrinsically drug resistant, making infections challenging to treat. The conserved whiB7 stress response is a key contributor to mycobacterial intrinsic drug resistance. Although we have a comprehensive structural and biochemical understanding of WhiB7, the complex set of signals that activate whiB7 expression remain less clear. It is believed that whiB7 expression is triggered by translational stalling in an upstream open reading frame (uORF) within the whiB7 5' leader, leading to antitermination and transcription into the downstream whiB7 ORF. To define the signals that activate whiB7, we employed a genome-wide CRISPRi epistasis screen and identified a diverse set of 150 mycobacterial genes whose inhibition results in constitutive whiB7 activation. Many of these genes encode amino acid biosynthetic enzymes, tRNAs, and tRNA synthetases, consistent with the proposed mechanism for whiB7 activation by translational stalling in the uORF. We show that the ability of the whiB7 5' regulatory region to sense amino acid starvation is determined by the coding sequence of the uORF. The uORF shows considerable sequence variation among different mycobacterial species, but it is universally and specifically enriched for alanine. Providing a potential rationalization for this enrichment, we find that while deprivation of many amino acids can activate whiB7 expression, whiB7 specifically coordinates an adaptive response to alanine starvation by engaging in a feedback loop with the alanine biosynthetic enzyme, aspC. Our results provide a holistic understanding of the biological pathways that influence whiB7 activation and reveal an extended role for the whiB7 pathway in mycobacterial physiology, beyond its canonical function in antibiotic resistance. These results have important implications for the design of combination drug treatments to avoid whiB7 activation, as well as help explain the conservation of this stress response across a wide range of pathogenic and environmental mycobacteria.
Figures
References
-
- World Health Organisation. WHO Global Tuberculosis Report 2021. (2021).
-
- Dick T. & Dartois V. TB drug susceptibility is more than MIC. Nat. Microbiol. 3, 971–972 (2018). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
