This is a preprint.
EZH2 inhibition promotes tumor immunogenicity in lung squamous cell carcinomas
- PMID: 37333199
- PMCID: PMC10274685
- DOI: 10.1101/2023.06.06.543919
EZH2 inhibition promotes tumor immunogenicity in lung squamous cell carcinomas
Update in
-
EZH2 Inhibition Promotes Tumor Immunogenicity in Lung Squamous Cell Carcinomas.Cancer Res Commun. 2024 Feb 13;4(2):388-403. doi: 10.1158/2767-9764.CRC-23-0399. Cancer Res Commun. 2024. PMID: 38265267 Free PMC article.
Abstract
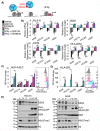
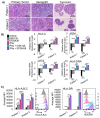
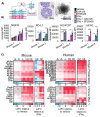
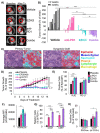
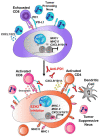
Two important factors that contribute to resistance to immune checkpoint inhibitors (ICIs) are an immune-suppressive microenvironment and limited antigen presentation by tumor cells. In this study, we examine if inhibition of the methyltransferase EZH2 can increase ICI response in lung squamous cell carcinomas (LSCCs). Our in vitro experiments using 2D human cancer cell lines as well as 3D murine and patient derived organoids treated with two inhibitors of the EZH2 plus interferon-γ (IFNγ) showed that EZH2 inhibition leads to expression of both major histocompatibility complex class I and II (MHCI/II) expression at both the mRNA and protein levels. ChIP-sequencing confirmed loss of EZH2-mediated histone marks and gain of activating histone marks at key loci. Further, we demonstrate strong tumor control in models of both autochthonous and syngeneic LSCC treated with anti-PD1 immunotherapy with EZH2 inhibition. Single-cell RNA sequencing and immune cell profiling demonstrated phenotypic changes towards more tumor suppressive phenotypes in EZH2 inhibitor treated tumors. These results indicate that this therapeutic modality could increase ICI responses in patients undergoing treatment for LSCC.
Keywords: EZH2; MHC; epigenetics; immunotherapy; neutrophils.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
