Whole genomes of deep-sea sponge-associated bacteria exhibit high novel natural product potential
- PMID: 37333438
- PMCID: PMC10117722
- DOI: 10.1093/femsmc/xtad005
Whole genomes of deep-sea sponge-associated bacteria exhibit high novel natural product potential
Abstract
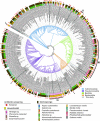
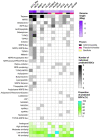
Global antimicrobial resistance is a health crisis that can change the face of modern medicine. Exploring diverse natural habitats for bacterially-derived novel antimicrobial compounds has historically been a successful strategy. The deep-sea presents an exciting opportunity for the cultivation of taxonomically novel organisms and exploring potentially chemically novel spaces. In this study, the draft genomes of 12 bacteria previously isolated from the deep-sea sponges Phenomena carpenteri and Hertwigia sp. are investigated for the diversity of specialized secondary metabolites. In addition, early data support the production of antibacterial inhibitory substances produced from a number of these strains, including activity against clinically relevant pathogens Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus. Draft whole-genomes are presented of 12 deep-sea isolates, which include four potentially novel strains: Psychrobacter sp. PP-21, Streptomyces sp. DK15, Dietzia sp. PP-33, and Micrococcus sp. M4NT. Across the 12 draft genomes, 138 biosynthetic gene clusters were detected, of which over half displayed less than 50% similarity to known BGCs, suggesting that these genomes present an exciting opportunity to elucidate novel secondary metabolites. Exploring bacterial isolates belonging to the phylum Actinomycetota, Pseudomonadota, and Bacillota from understudied deep-sea sponges provided opportunities to search for new chemical diversity of interest to those working in antibiotic discovery.
Keywords: antibiotic discovery; bacterial genomics; biosynthetic gene clusters; deep sea; natural products; porifera.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Impact of growth media and pressure on the diversity and antimicrobial activity of isolates from two species of hexactinellid sponge.Microbiology (Reading). 2021 Dec;167(12):001123. doi: 10.1099/mic.0.001123. Microbiology (Reading). 2021. PMID: 34898418 Free PMC article.
-
Diverse and Abundant Secondary Metabolism Biosynthetic Gene Clusters in the Genomes of Marine Sponge Derived Streptomyces spp. Isolates.Mar Drugs. 2018 Feb 20;16(2):67. doi: 10.3390/md16020067. Mar Drugs. 2018. PMID: 29461500 Free PMC article.
-
Biosynthetic gene cluster signature profiles of pathogenic Gram-negative bacteria isolated from Egyptian clinical settings.Microbiol Spectr. 2023 Sep 14;11(5):e0134423. doi: 10.1128/spectrum.01344-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37707241 Free PMC article.
-
Comparative Metagenomic Analysis of Biosynthetic Diversity across Sponge Microbiomes Highlights Metabolic Novelty, Conservation, and Diversification.mSystems. 2022 Aug 30;7(4):e0035722. doi: 10.1128/msystems.00357-22. Epub 2022 Jul 18. mSystems. 2022. PMID: 35862823 Free PMC article. Review.
-
A comprehensive review of marine sponge metabolites, with emphasis on Neopetrosia sp.Int J Biol Macromol. 2024 Nov;280(Pt 2):135823. doi: 10.1016/j.ijbiomac.2024.135823. Epub 2024 Sep 21. Int J Biol Macromol. 2024. PMID: 39313052 Review.
References
-
- Abdel-Mageed WM, Lehri B, Jarmusch SAet al. . Whole genome sequencing of four bacterial strains from South Shetland Trench revealing biosynthetic and environmental adaptation gene clusters. Mar Geonomics. 2020;54:100782. - PubMed
-
- Balitz DM, Bush JA, Bradner WTet al. . Isolation of Lavendamycin a new antibiotic from Streptomyces lavendulae. J Antibiot (Tokyo). 1982;35:259–65. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
