PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
- PMID: 37333450
- PMCID: PMC10276617
- DOI: 10.1515/med-2023-0727
PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
Abstract
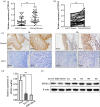
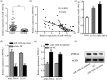
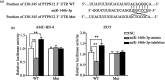
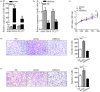
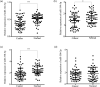
Laryngeal squamous cell carcinoma (LSCC) is a common malignancy among men in the anatomical position of head and neck. Hoarseness, pharyngalgia, and dyspnea are common symptoms. LSCC is a complex polygenic carcinoma that is caused by many factors involving polygenic alteration, environmental pollution, tobacco, and human papillomavirus. Classical protein tyrosine phosphatase nonreceptor type 12 (PTPN12) has been extensively studied to decipher its mechanism as a tumor suppressor gene in various human carcinomas; however, there is no comprehensive elucidation of the PTPN12 expression and its regulatory mechanisms in LSCC. As such, we expect to provide new insights for finding new biomarkers and effective therapeutic targets in LSCC. Immunohistochemical staining, western blot (WB), and quantitative real-time RT-PCR (qRT-PCR) were used for the messenger RNA (mRNA) and protein expression analyses of PTPN12, respectively. 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, clone formation, transwell migration, and transwell invasion assays were used to assess the proliferation, migration, and invasion ability of LSCC cells. Online prediction and design software tools (http://www.targetscan.org/ and http://www.microRNA.org) were used to predict associated miRNA. Studying the targeted regulatory relationship between miR-146b-3p and PTPN12 was based on dual luciferase reporter gene analysis. qRT-PCR was used to assess miR-146b-3p expression in LSCC. miR-146b-3p inhibitor and mimic were transfected, followed by qRT-PCR and WB assays to detect the expression of PTPN12. The gain and loss functional experiments were used to investigate the effects of miR-146b-3p transfection on the proliferation, migration, and invasion of tumor cells. Online bioinformatics prediction software (https://cn.string-db.org/ and https://www.genecards.org/) was used to determine potential downstream target genes of PTPN12. qRT-PCR and WB analyses were used to assess the mRNA and protein expression levels of target genes. Our study showed significantly decreased mRNA and protein expression levels of PTPN12 in LSCC compared with the adjacent normal tissues. The lower PTPN12 mRNA expression was correlated with pathological differentiation, and lower PTPN12 protein expression was correlated with the TNM stage in LSCC tissues. The subsequent in vitro functional analyses showed the inhibitory effect of PTPN12 over-expression on the proliferation, migration, and invasiveness abilities of LSCC cell line. Using online prediction and design software, miR-146b-3p was searched to target PTPN12. The miR-146b-3p was expressed at a high level in LSCC tissues and cell lines. Luciferase reporter assay exhibited that miR-146b-3p inhibited the luciferase activity of PTPN12 markedly. The functional analyses showed the tumor-promoting role of miR-146b-3p on the proliferation, migration, and invasiveness abilities of LSCC cell. Furthermore, co-transfection of cells with miR-146b-3p and PTPN12 significantly restored the inhibitory effect of PTPN12 on LSCC cell growth, migration, and invasiveness. This phenomenon unveiled that miR-146b-3p regulated the proliferation, migration, and invasion of LSCC cells by targeting PTPN12. EGFR and ERBB2 were selected as the downstream-regulation target genes. Up-regulation of PTPN12 significantly suppressed EGFR expression. Accordingly, the miR-146b-3p mimic significantly up-regulated the EGFR expression. However, up-regulation of PTPN12 and miR-146b-3p mimic suppressed ERBB2 protein expression but induced its gene expression. Down-regulation of PTPN12 is associated with up-regulation of miR-146b-3p in LSCC. Moreover, PTPN12 serves as a tumor suppressor gene through regulating the proliferation, migration, and invasion of LSCC cells. miR-146b-3p/PTPN12 axis is expected to be a novel therapeutic target in LSCC.
Keywords: Laryngeal squamous cell carcinoma; PTPN12; invasion; miR-146b-3p; migration.
© 2023 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
