The role of individual exopolysaccharides in antibiotic tolerance of Pseudomonas aeruginosa aggregates
- PMID: 37333638
- PMCID: PMC10272609
- DOI: 10.3389/fmicb.2023.1187708
The role of individual exopolysaccharides in antibiotic tolerance of Pseudomonas aeruginosa aggregates
Abstract
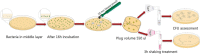
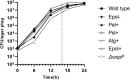
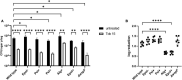
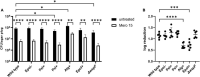
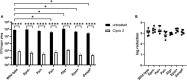
The bacterium Pseudomonas aeruginosa is involved in chronic infections of cystic fibrosis lungs and chronic wounds. In these infections the bacteria are present as aggregates suspended in host secretions. During the course of the infections there is a selection for mutants that overproduce exopolysaccharides, suggesting that the exopolysaccharides play a role in the persistence and antibiotic tolerance of the aggregated bacteria. Here, we investigated the role of individual P. aeruginosa exopolysaccharides in aggregate-associated antibiotic tolerance. We employed an aggregate-based antibiotic tolerance assay on a set of P. aeruginosa strains that were genetically engineered to over-produce a single, none, or all of the three exopolysaccharides Pel, Psl, and alginate. The antibiotic tolerance assays were conducted with the clinically relevant antibiotics tobramycin, ciprofloxacin and meropenem. Our study suggests that alginate plays a role in the tolerance of P. aeruginosa aggregates toward tobramycin and meropenem, but not ciprofloxacin. However, contrary to previous studies we did not observe a role for Psl or Pel in the tolerance of P. aeruginosa aggregates toward tobramycin, ciprofloxacin, and meropenem.
Keywords: Pseudomonas aeruginosa; aggregates; antibiotic tolerance; biofilm; extracellular matrix.
Copyright © 2023 Liang, Nilsson, Kragh, Hedal, Alcàcer-Almansa, Kiilerich, Andersen and Tolker-Nielsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

