Whole-genome sequencing analysis of Klebsiella aerogenes among men who have sex with men in Guangzhou, China
- PMID: 37333657
- PMCID: PMC10272549
- DOI: 10.3389/fmicb.2023.1102907
Whole-genome sequencing analysis of Klebsiella aerogenes among men who have sex with men in Guangzhou, China
Abstract
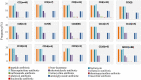
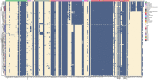
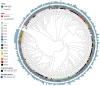
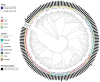
Klebsiella aerogenes is a common infectious bacterium that poses a threat to human health. Nevertheless, there are limited data on the population structure, genetic diversity, and pathogenicity of K. aerogenes, especially among men who have sex with men (MSM). The present study aimed to clarify the sequence types (STs), clonal complexes (CCs), resistance genes, and virulence factors of popular strains. Multilocus sequence typing was used to describe the population structure of K. aerogenes. The Virulence Factor Database and Comprehensive Antibiotic Resistance Database were used to assess the virulence and resistance profiles. In this study, next-generation sequencing was performed on nasal swabs specimens collected in an HIV Voluntary Counseling Testing outpatient department in Guangzhou, China, from April to August 2019. The identification results showed that a total of 258 K. aerogenes isolates were collected from 911 participants. We found that the isolates were most resistant to furantoin (89.53%, 231/258) and ampicillin (89.15%, 230/258), followed by imipenem (24.81%, 64/258) and cefotaxime (18.22%, 47/258). The most common STs in carbapenem-resistant K. aerogenes were ST4, ST93, and ST14. The population has at least 14 CCs, including several novel ones identified in this study (CC11-CC16). The main mechanism of drug resistance genes was antibiotic efflux. Based on the presence of the iron carrier production genes irp and ybt, we identified two clusters according to virulence profiles. In cluster A, CC3 and CC4 carry the clb operator encoding the toxin. Increased monitoring is needed for the three main ST type strains carried by MSM. The main clone group CC4 has a large number of toxin genes, and it spreads among MSM. Caution is needed to prevent further spread of this clone group in this population. In sum, our results may provide a foundation for the development of new therapeutic and surveillance strategies for treating MSM.
Keywords: Enterobacteriaceae; antimicrobial resistance; multidrug-resistant (MDR); pathogens; whole genome sequencing (WGS).
Copyright © 2023 Cheng, Ma, Gong, Liang, Guo, Ye, Han and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Bioinformatics, Babraham Institute.
-
- Biendo M., Canarelli B., Thomas D., Rousseau F., Hamdad F., Adjide C., et al. (2008). Successive emergence of extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacter aerogenes isolates in a university hospital. J. Clin. Microbiol. 46, 1037–1044. 10.1128/JCM.00197-07 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

