CNN-based automatic segmentations and radiomics feature reliability on contrast-enhanced ultrasound images for renal tumors
- PMID: 37333811
- PMCID: PMC10272725
- DOI: 10.3389/fonc.2023.1166988
CNN-based automatic segmentations and radiomics feature reliability on contrast-enhanced ultrasound images for renal tumors
Abstract
Objective: To investigate the feasibility and efficiency of automatic segmentation of contrast-enhanced ultrasound (CEUS) images in renal tumors by convolutional neural network (CNN) based models and their further application in radiomic analysis.
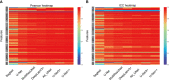
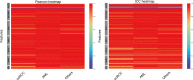
Materials and methods: From 94 pathologically confirmed renal tumor cases, 3355 CEUS images were extracted and randomly divided into training set (3020 images) and test set (335 images). According to the histological subtypes of renal cell carcinoma, the test set was further split into clear cell renal cell carcinoma (ccRCC) set (225 images), renal angiomyolipoma (AML) set (77 images) and set of other subtypes (33 images). Manual segmentation was the gold standard and serves as ground truth. Seven CNN-based models including DeepLabV3+, UNet, UNet++, UNet3+, SegNet, MultilResUNet and Attention UNet were used for automatic segmentation. Python 3.7.0 and Pyradiomics package 3.0.1 were used for radiomic feature extraction. Performance of all approaches was evaluated by the metrics of mean intersection over union (mIOU), dice similarity coefficient (DSC), precision, and recall. Reliability and reproducibility of radiomics features were evaluated by the Pearson coefficient and the intraclass correlation coefficient (ICC).
Results: All seven CNN-based models achieved good performance with the mIOU, DSC, precision and recall ranging between 81.97%-93.04%, 78.67%-92.70%, 93.92%-97.56%, and 85.29%-95.17%, respectively. The average Pearson coefficients ranged from 0.81 to 0.95, and the average ICCs ranged from 0.77 to 0.92. The UNet++ model showed the best performance with the mIOU, DSC, precision and recall of 93.04%, 92.70%, 97.43% and 95.17%, respectively. For ccRCC, AML and other subtypes, the reliability and reproducibility of radiomic analysis derived from automatically segmented CEUS images were excellent, with the average Pearson coefficients of 0.95, 0.96 and 0.96, and the average ICCs for different subtypes were 0.91, 0.93 and 0.94, respectively.
Conclusion: This retrospective single-center study showed that the CNN-based models had good performance on automatic segmentation of CEUS images for renal tumors, especially the UNet++ model. The radiomics features extracted from automatically segmented CEUS images were feasible and reliable, and further validation by multi-center research is necessary.
Keywords: UNet; automatic segmentation; contrast-enhanced ultrasound; deep learning; renal tumors.
Copyright © 2023 Yang, Chen, Liang, Bai, Wang, Zhao, Ma, Niu, Li, Xie and Cai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Multiple U-Net-Based Automatic Segmentations and Radiomics Feature Stability on Ultrasound Images for Patients With Ovarian Cancer.Front Oncol. 2021 Feb 18;10:614201. doi: 10.3389/fonc.2020.614201. eCollection 2020. Front Oncol. 2021. PMID: 33680934 Free PMC article.
-
Comparative analysis of deep learning and radiomic signatures for overall survival prediction in recurrent high-grade glioma treated with immunotherapy.Cancer Imaging. 2025 Jan 21;25(1):5. doi: 10.1186/s40644-024-00818-0. Cancer Imaging. 2025. PMID: 39838503 Free PMC article.
-
The Accuracy and Radiomics Feature Effects of Multiple U-net-Based Automatic Segmentation Models for Transvaginal Ultrasound Images of Cervical Cancer.J Digit Imaging. 2022 Aug;35(4):983-992. doi: 10.1007/s10278-022-00620-z. Epub 2022 Mar 30. J Digit Imaging. 2022. PMID: 35355160 Free PMC article. Clinical Trial.
-
A deep-learning approach for segmentation of liver tumors in magnetic resonance imaging using UNet+.BMC Cancer. 2023 Nov 3;23(1):1060. doi: 10.1186/s12885-023-11432-x. BMC Cancer. 2023. PMID: 37923988 Free PMC article.
-
Deep learning-based recognition and segmentation of intracranial aneurysms under small sample size.Front Physiol. 2022 Dec 19;13:1084202. doi: 10.3389/fphys.2022.1084202. eCollection 2022. Front Physiol. 2022. PMID: 36601346 Free PMC article.
Cited by
-
Segmentation of glioblastomas via 3D FusionNet.Front Oncol. 2024 Nov 15;14:1488616. doi: 10.3389/fonc.2024.1488616. eCollection 2024. Front Oncol. 2024. PMID: 39619438 Free PMC article.
-
Artificial Intelligence-Augmented Advancements in the Diagnostic Challenges Within Renal Cell Carcinoma.J Clin Med. 2025 Mar 26;14(7):2272. doi: 10.3390/jcm14072272. J Clin Med. 2025. PMID: 40217721 Free PMC article. Review.
-
Automation of Wilms' tumor segmentation by artificial intelligence.Cancer Imaging. 2024 Jul 2;24(1):83. doi: 10.1186/s40644-024-00729-0. Cancer Imaging. 2024. PMID: 38956718 Free PMC article.
-
The impact of BTK knockdown on lung adenocarcinoma growth and immune response.Cancer Sci. 2025 Jun;116(6):1550-1564. doi: 10.1111/cas.16394. Epub 2024 Dec 4. Cancer Sci. 2025. PMID: 39628301 Free PMC article.
-
Ultrasound-based radiogenomics: status, applications, and future direction.Ultrasonography. 2025 Mar;44(2):95-111. doi: 10.14366/usg.24152. Epub 2024 Dec 12. Ultrasonography. 2025. PMID: 39935290 Free PMC article.
References
LinkOut - more resources
Full Text Sources

