Exploring personalized structural connectomics for moderate to severe traumatic brain injury
- PMID: 37334004
- PMCID: PMC10270710
- DOI: 10.1162/netn_a_00277
Exploring personalized structural connectomics for moderate to severe traumatic brain injury
Abstract
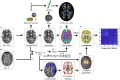
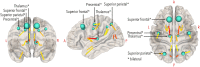
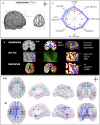
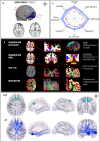
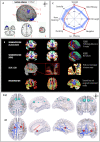
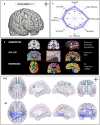
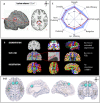
Graph theoretical analysis of the structural connectome has been employed successfully to characterize brain network alterations in patients with traumatic brain injury (TBI). However, heterogeneity in neuropathology is a well-known issue in the TBI population, such that group comparisons of patients against controls are confounded by within-group variability. Recently, novel single-subject profiling approaches have been developed to capture inter-patient heterogeneity. We present a personalized connectomics approach that examines structural brain alterations in five chronic patients with moderate to severe TBI who underwent anatomical and diffusion magnetic resonance imaging. We generated individualized profiles of lesion characteristics and network measures (including personalized graph metric GraphMe plots, and nodal and edge-based brain network alterations) and compared them against healthy reference cases (N = 12) to assess brain damage qualitatively and quantitatively at the individual level. Our findings revealed alterations of brain networks with high variability between patients. With validation and comparison to stratified, normative healthy control comparison cohorts, this approach could be used by clinicians to formulate a neuroscience-guided integrative rehabilitation program for TBI patients, and for designing personalized rehabilitation protocols based on their unique lesion load and connectome.
Keywords: Graph theory; Lesion filling; Personalized connectomics; Personalized medicine; Structural connectomics; Traumatic brain injury.
© 2022 Massachusetts Institute of Technology.
Figures
References
-
- Avants, B. B., Tustison, N., & Song, G. (2009). Advanced normalization tools (ANTS). Insight Journal, 2(365), 1–35. 10.54294/uvnhin - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
