Variation in genomic traits of microbial communities among ecosystems
- PMID: 37334231
- PMCID: PMC10117780
- DOI: 10.1093/femsmc/xtab020
Variation in genomic traits of microbial communities among ecosystems
Abstract
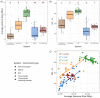
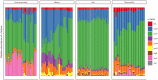
Free-living bacteria in nutrient limited environments often exhibit traits which may reduce the cost of reproduction, such as smaller genome size, low GC content and fewer sigma (σ) factor and 16S rRNA gene copies. Despite the potential utility of these traits to detect relationships between microbial communities and ecosystem-scale properties, few studies have assessed these traits on a community-scale. Here, we analysed these traits from publicly available metagenomes derived from marine, soil, host-associated and thermophilic communities. In marine and thermophilic communities, genome size and GC content declined in parallel, consistent with genomic streamlining, with GC content in thermophilic communities generally higher than in marine systems. In contrast, soil communities averaging smaller genomes featured higher GC content and were often from low-carbon environments, suggesting unique selection pressures in soil bacteria. The abundance of specific σ-factors varied with average genome size and ecosystem type. In oceans, abundance of fliA, a σ-factor controlling flagella biosynthesis, was positively correlated with community average genome size-reflecting known trade-offs between nutrient conservation and chemotaxis. In soils, a high abundance of the stress response σ-factor gene rpoS was associated with smaller average genome size and often located in harsh and/or carbon-limited environments-a result which tracks features observed in culture and indicates an increased capacity for stress response in nutrient-poor soils. This work shows how ecosystem-specific constraints are associated with trade-offs which are embedded in the genomic features of bacteria in microbial communities, and which can be detected at the community level, highlighting the importance of genomic features in microbial community analysis.
Keywords: GC content; genome size; metagenomics; sigma-factors; soil; streamlining.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures



References
-
- Barberán A, Ramirez KS, Leff JWet al. . Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett. 2014;17:794–802. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
