Emergence of resistance-associated variants during sofosbuvir treatment in chronically infected hepatitis E patients
- PMID: 37334496
- PMCID: PMC10653298
- DOI: 10.1097/HEP.0000000000000514
Emergence of resistance-associated variants during sofosbuvir treatment in chronically infected hepatitis E patients
Abstract
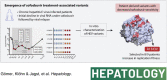
Background and aims: Chronic HEV infections remain a serious problem in immunocompromised patients, as specifically approved antiviral drugs are unavailable. In 2020, a 24-week multicenter phase II pilot trial was carried out, evaluating the nucleotide analog sofosbuvir by treating nine chronically HEV-infected patients with sofosbuvir (Trial Number NCT03282474). During the study, antiviral therapy reduced virus RNA levels initially but did not lead to a sustained virologic response. Here, we characterize the changes in HEV intrahost populations during sofosbuvir treatment to identify the emergence of treatment-associated variants.
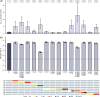
Approach and results: We performed high-throughput sequencing on RNA-dependent RNA polymerase sequences to characterize viral population dynamics in study participants. Subsequently, we used an HEV-based reporter replicon system to investigate sofosbuvir sensitivity in high-frequency variants. Most patients had heterogenous HEV populations, suggesting high adaptability to treatment-related selection pressures. We identified numerous amino acid alterations emerging during treatment and found that the EC 50 of patient-derived replicon constructs was up to ~12-fold higher than the wild-type control, suggesting that variants associated with lower drug sensitivity were selected during sofosbuvir treatment. In particular, a single amino acid substitution (A1343V) in the finger domain of ORF1 could reduce susceptibility to sofosbuvir significantly in 8 of 9 patients.
Conclusions: In conclusion, viral population dynamics played a critical role during antiviral treatment. High population diversity during sofosbuvir treatment led to the selection of variants (especially A1343V) with lower sensitivity to the drug, uncovering a novel mechanism of resistance-associated variants during sofosbuvir treatment.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
Thomas Horvatits is on the speakers’ bureau for Microgen diagnostics. Julian Schulze zur Wiesch is on the speakers‘ bureau for Gilead. Markus Cornberg advises and is on the speakers’ bureau for AbbVie, Gilead, and MSD. He advises AiCuris, GlaxoSmithKline, Jansen-Cilag, Novartis, Roche, and Swedish Orphan Biovitrum. He is on the speakers’ bureau for Falk. Heiner Wedemeyer consults, is on the speakers’ bureau, received grants, and has other interests with Gilead. He consults, received grants, and has other interests with Falk. He consults and has other interests with MSD. He consults and advises Roche. He consults and is on the speakers’ bureau for Pfizer. He is on the speakers’ bureau for the Falk Foundation. The remaining authors have no conflicts to report.
Figures




Comment in
-
Toward antivirals against hepatitis E: In the steps of hepatitis C.Hepatology. 2023 Dec 1;78(6):1695-1697. doi: 10.1097/HEP.0000000000000509. Epub 2023 Jun 12. Hepatology. 2023. PMID: 37300387 No abstract available.
References
-
- World Health Organization. Hepatitis E Fact Sheets [Internet]. 2022 [Accessed cited 2023 Jan 3]; https://www.who.int/news-room/fact-sheets/detail/hepatitis-e
-
- Velavan TP, Pallerla SR, Johne R, Todt D, Steinmann E, Schemmerer M, et al. Hepatitis E: An update on One Health and clinical medicine. Liver Int. 2021;41:1462–1473. - PubMed
-
- Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of Hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361:1025–1027. - PubMed
-
- Singh GKJ, Ijaz S, Rockwood N, Farnworth SP, Devitt E, Atkins M, et al. Chronic Hepatitis E as a cause for cryptogenic cirrhosis in HIV. J Infect. 2013;66:103–106. - PubMed
-
- Gérolami R, Moal V, Colson P. Chronic Hepatitis E with cirrhosis in a kidney-transplant recipient. N Engl J Med. 2008;358:859–860. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

