The role of aging and brain-derived neurotrophic factor signaling in expression of base excision repair genes in the human brain
- PMID: 37334527
- PMCID: PMC10497833
- DOI: 10.1111/acel.13905
The role of aging and brain-derived neurotrophic factor signaling in expression of base excision repair genes in the human brain
Abstract
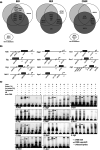
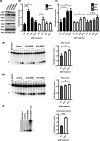
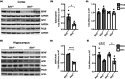
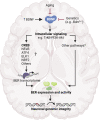
DNA damage is a central contributor to the aging process. In the brain, a major threat to the DNA is the considerable amount of reactive oxygen species produced, which can inflict oxidative DNA damage. This type of damage is removed by the base excision repair (BER) pathway, an essential DNA repair mechanism, which contributes to genome stability in the brain. Despite the crucial role of the BER pathway, insights into how this pathway is affected by aging in the human brain and the underlying regulatory mechanisms are very limited. By microarray analysis of four cortical brain regions from humans aged 20-99 years (n = 57), we show that the expression of core BER genes is largely downregulated during aging across brain regions. Moreover, we find that expression of many BER genes correlates positively with the expression of the neurotrophin brain-derived neurotrophic factor (BDNF) in the human brain. In line with this, we identify binding sites for the BDNF-activated transcription factor, cyclic-AMP response element-binding protein (CREB), in the promoter of most BER genes and confirm the ability of BDNF to regulate several BER genes by BDNF treatment of mouse primary hippocampal neurons. Together, these findings uncover the transcriptional landscape of BER genes during aging of the brain and suggest BDNF as an important regulator of BER in the human brain.
Keywords: DNA repair; aging; base excision repair; brain; brain-derived neurotrophic factor; cyclic-AMP response element-binding protein; neurons.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Base excision repair in the mammalian brain: implication for age related neurodegeneration.Mech Ageing Dev. 2013 Oct;134(10):440-8. doi: 10.1016/j.mad.2013.04.005. Epub 2013 May 2. Mech Ageing Dev. 2013. PMID: 23643943 Free PMC article.
-
Differential Regulation of the BDNF Gene in Cortical and Hippocampal Neurons.J Neurosci. 2022 Dec 7;42(49):9110-9128. doi: 10.1523/JNEUROSCI.2535-21.2022. Epub 2022 Oct 31. J Neurosci. 2022. PMID: 36316156 Free PMC article.
-
Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I.J Biol Chem. 2002 Sep 27;277(39):35920-31. doi: 10.1074/jbc.M204784200. Epub 2002 Jul 11. J Biol Chem. 2002. PMID: 12114522
-
DNA repair in neurons: so if they don't divide what's to repair?Mutat Res. 2007 Jan 3;614(1-2):24-36. doi: 10.1016/j.mrfmmm.2006.06.007. Epub 2006 Aug 1. Mutat Res. 2007. PMID: 16879837 Review.
-
DNA repair, mitochondria, and neurodegeneration.Neuroscience. 2007 Apr 14;145(4):1318-29. doi: 10.1016/j.neuroscience.2006.08.061. Epub 2006 Nov 7. Neuroscience. 2007. PMID: 17092652 Review.
Cited by
-
Age, cancer, and the dual burden of cancer and doxorubicin in skeletal muscle wasting in female rats: which one to blame?Biogerontology. 2025 Jan 24;26(1):47. doi: 10.1007/s10522-024-10182-y. Biogerontology. 2025. PMID: 39853446
-
Harnessing BDNF Signaling to Promote Resilience in Aging.Aging Dis. 2024 Sep 3;16(4):1813-1841. doi: 10.14336/AD.2024.0961. Aging Dis. 2024. PMID: 39656488 Free PMC article. Review.
-
Proteins Associated with Neurodegenerative Diseases: Link to DNA Repair.Biomedicines. 2024 Dec 11;12(12):2808. doi: 10.3390/biomedicines12122808. Biomedicines. 2024. PMID: 39767715 Free PMC article. Review.
-
Markers of Mitochondrial Function and DNA Repair Associated with Physical Function in Centenarians.Biomolecules. 2024 Jul 26;14(8):909. doi: 10.3390/biom14080909. Biomolecules. 2024. PMID: 39199297 Free PMC article.
References
-
- Aamann, M. D. , Hvitby, C. , Popuri, V. , Muftuoglu, M. , Lemminger, L. , Skeby, C. K. , Keijzers, G. , Ahn, B. , Bjoras, M. , Bohr, V. A. , & Stevnsner, T. (2014). Cockayne syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mechanisms of Ageing and Development, 135, 1–14. - PMC - PubMed
-
- Alexeeva, M. , Moen, M. N. , Xu, X. M. , Rasmussen, A. , Leiros, I. , Kirpekar, F. , Klungland, A. , Alsøe, L. , Nilsen, H. , & Bjelland, S. (2021). Intrinsic Strand‐incision activity of human UNG: Implications for Nick generation in immunoglobulin gene diversification. Frontiers in Immunology, 12, 762032. - PMC - PubMed
-
- Ayala‐Torres, S. , Chen, Y. , Svoboda, T. , Rosenblatt, J. , & Van Houten, B. (2000). Analysis of gene‐specific DNA damage and repair using quantitative polymerase chain reaction. Methods, 22(2), 135–147. - PubMed

