In skeletal muscle and neural crest cells, SMCHD1 regulates biological pathways relevant for Bosma syndrome and facioscapulohumeral dystrophy phenotype
- PMID: 37334829
- PMCID: PMC10415154
- DOI: 10.1093/nar/gkad523
In skeletal muscle and neural crest cells, SMCHD1 regulates biological pathways relevant for Bosma syndrome and facioscapulohumeral dystrophy phenotype
Abstract
Many genetic syndromes are linked to mutations in genes encoding factors that guide chromatin organization. Among them, several distinct rare genetic diseases are linked to mutations in SMCHD1 that encodes the structural maintenance of chromosomes flexible hinge domain containing 1 chromatin-associated factor. In humans, its function as well as the impact of its mutations remains poorly defined. To fill this gap, we determined the episignature associated with heterozygous SMCHD1 variants in primary cells and cell lineages derived from induced pluripotent stem cells for Bosma arhinia and microphthalmia syndrome (BAMS) and type 2 facioscapulohumeral dystrophy (FSHD2). In human tissues, SMCHD1 regulates the distribution of methylated CpGs, H3K27 trimethylation and CTCF at repressed chromatin but also at euchromatin. Based on the exploration of tissues affected either in FSHD or in BAMS, i.e. skeletal muscle fibers and neural crest stem cells, respectively, our results emphasize multiple functions for SMCHD1, in chromatin compaction, chromatin insulation and gene regulation with variable targets or phenotypical outcomes. We concluded that in rare genetic diseases, SMCHD1 variants impact gene expression in two ways: (i) by changing the chromatin context at a number of euchromatin loci or (ii) by directly regulating some loci encoding master transcription factors required for cell fate determination and tissue differentiation.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

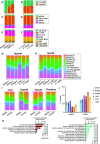
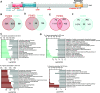
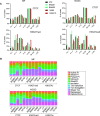



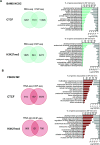
References
-
- Chen K., Czabotar P.E., Blewitt M.E., Murphy J.M.. The hinge domain of the epigenetic repressor Smchd1 adopts an unconventional homodimeric configuration. Biochem. J. 2016; 473:733–742. - PubMed
-
- Gendrel A.V., Apedaile A., Coker H., Termanis A., Zvetkova I., Godwin J., Tang Y.A., Huntley D., Montana G., Taylor S.et al.. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell. 2012; 23:265–279. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

