All these screens that we've done: how functional genetic screens have informed our understanding of ribosome biogenesis
- PMID: 37335083
- PMCID: PMC10329186
- DOI: 10.1042/BSR20230631
All these screens that we've done: how functional genetic screens have informed our understanding of ribosome biogenesis
Abstract
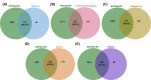
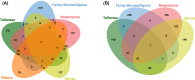
Ribosome biogenesis is the complex and essential process that ultimately leads to the synthesis of cellular proteins. Understanding each step of this essential process is imperative to increase our understanding of basic biology, but also more critically, to provide novel therapeutic avenues for genetic and developmental diseases such as ribosomopathies and cancers which can arise when this process is impaired. In recent years, significant advances in technology have made identifying and characterizing novel human regulators of ribosome biogenesis via high-content, high-throughput screens. Additionally, screening platforms have been used to discover novel therapeutics for cancer. These screens have uncovered a wealth of knowledge regarding novel proteins involved in human ribosome biogenesis, from the regulation of the transcription of the ribosomal RNA to global protein synthesis. Specifically, comparing the discovered proteins in these screens showed interesting connections between large ribosomal subunit (LSU) maturation factors and earlier steps in ribosome biogenesis, as well as overall nucleolar integrity. In this review, a discussion of the current standing of screens for human ribosome biogenesis factors through the lens of comparing the datasets and discussing the biological implications of the areas of overlap will be combined with a look toward other technologies and how they can be adapted to discover more factors involved in ribosome synthesis, and answer other outstanding questions in the field.
Keywords: RNA; high-throughput screening; nucleolus; ribosome biogenesis; ribosomes.
© 2023 The Author(s).
Conflict of interest statement
The author declares that there are no competing interests associated with the manuscript.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

