Analysis of the potential relationship between COVID-19 and Behcet's disease using transcriptome data
- PMID: 37335738
- PMCID: PMC10193850
- DOI: 10.1097/MD.0000000000033821
Analysis of the potential relationship between COVID-19 and Behcet's disease using transcriptome data
Abstract
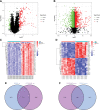
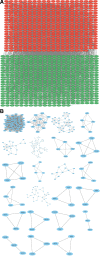
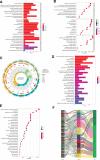
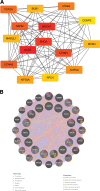
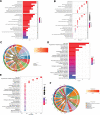
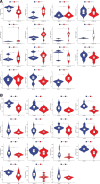
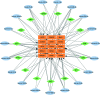
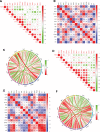
To investigate the potential role of COVID-19 in relation to Behcet's disease (BD) and to search for relevant biomarkers. We used a bioinformatics approach to download transcriptomic data from peripheral blood mononuclear cells (PBMCs) of COVID-19 patients and PBMCs of BD patients, screened the common differential genes between COVID-19 and BD, performed gene ontology (GO) and pathway analysis, and constructed the protein-protein interaction (PPI) network, screened the hub genes and performed co-expression analysis. In addition, we constructed the genes-transcription factors (TFs)-miRNAs network, the genes-diseases network and the genes-drugs network to gain insight into the interactions between the 2 diseases. We used the RNA-seq dataset from the GEO database (GSE152418, GSE198533). We used cross-analysis to obtain 461 up-regulated common differential genes and 509 down-regulated common differential genes, mapped the PPI network, and used Cytohubba to identify the 15 most strongly associated genes as hub genes (ACTB, BRCA1, RHOA, CCNB1, ASPM, CCNA2, TOP2A, PCNA, AURKA, KIF20A, MAD2L1, MCM4, BUB1, RFC4, and CENPE). We screened for statistically significant hub genes and found that ACTB was in low expression of both BD and COVID-19, and ASPM, CCNA2, CCNB1, and CENPE were in low expression of BD and high expression of COVID-19. GO analysis and pathway analysis was then performed to obtain common pathways and biological response processes, which suggested a common association between BD and COVID-19. The genes-TFs-miRNAs network, genes-diseases network and genes-drugs network also play important roles in the interaction between the 2 diseases. Interaction between COVID-19 and BD exists. ACTB, ASPM, CCNA2, CCNB1, and CENPE as potential biomarkers for 2 diseases.
Copyright © 2023 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

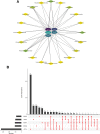
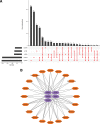

References
-
- Bulur I, Onder M. Behcet disease: new aspects. Clin Dermatol. 2017;35:421–34. - PubMed
-
- Shimizu J, Murayama MA, Miyabe Y, et al. Immunopathology of Behcet’s disease: an overview of the metagenomic approaches. Rheumato. 2022;2:74–86.
-
- Dincses E, Esatoglu S, Fresko I, et al. Outcome of invasive procedures for venous thrombosis in Behçet’s syndrome: case series and systematic literature review. Clin Exp Rheumatol. 2019:125–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

