Dynamical scattering in ice-embedded proteins in conventional and scanning transmission electron microscopy
- PMID: 37335769
- PMCID: PMC10324487
- DOI: 10.1107/S2052252523004505
Dynamical scattering in ice-embedded proteins in conventional and scanning transmission electron microscopy
Abstract
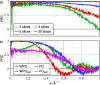
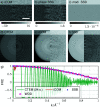
Structure determination of biological macromolecules using cryogenic electron microscopy is based on applying the phase object (PO) assumption and the weak phase object (WPO) approximation to reconstruct the 3D potential density of the molecule. To enhance the understanding of image formation of protein complexes embedded in glass-like ice in a transmission electron microscope, this study addresses multiple scattering in tobacco mosaic virus (TMV) specimens. This includes the propagation inside the molecule while also accounting for the effect of structural noise. The atoms in biological macromolecules are light but are distributed over several nanometres. Commonly, PO and WPO approximations are used in most simulations and reconstruction models. Therefore, dynamical multislice simulations of TMV specimens embedded in glass-like ice were performed based on fully atomistic molecular-dynamics simulations. In the first part, the impact of multiple scattering is studied using different numbers of slices. In the second part, different sample thicknesses of the ice-embedded TMV are considered in terms of additional ice layers. It is found that single-slice models yield full frequency transfer up to a resolution of 2.5 Å, followed by attenuation up to 1.4 Å. Three slices are sufficient to reach an information transfer up to 1.0 Å. In the third part, ptychographic reconstructions based on scanning transmission electron microscopy (STEM) and single-slice models are compared with conventional TEM simulations. The ptychographic reconstructions do not need the deliberate introduction of aberrations, are capable of post-acquisition aberration correction and promise benefits for information transfer, especially at resolutions beyond 1.8 Å.
Keywords: amorphous ice; cryogenic electron microscopy; dynamical scattering; image simulations; integrative structural biology; molecular dynamics.
open access.
Figures







References
-
- Abascal, J. L. & Vega, C. (2005). J. Chem. Phys. 123, 234505. - PubMed
-
- Angert, I., Burmester, C., Dinges, C., Rose, H. & Schröder, R. (1996). Ultramicroscopy, 63, 181–192.
-
- Bangun, A., Melnyk, O., März, B., Diederichs, B., Clausen, A., Weber, D., Filbir, F. & Müller-Caspary, K. (2022). IEEE Trans. Comput. Imaging, 8, 996–1011.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

